N-Linked glycosylation of the membrane protein ectodomain regulates infectious bronchitis virus-induced ER stress response, apoptosis and pathogenesis
- PMID: 30852271
- PMCID: PMC7112112
- DOI: 10.1016/j.virol.2019.02.017
N-Linked glycosylation of the membrane protein ectodomain regulates infectious bronchitis virus-induced ER stress response, apoptosis and pathogenesis
Abstract
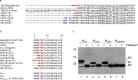
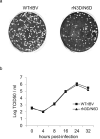
Coronavirus membrane (M) protein is the most abundant structural protein playing a critical role in virion assembly. Previous studies show that the N-terminal ectodomain of M protein is modified by glycosylation, but its precise functions are yet to be thoroughly investigated. In this study, we confirm that N-linked glycosylation occurs at two predicted sites in the M protein ectodomain of infectious bronchitis coronavirus (IBV). Dual mutations at the two sites (N3D/N6D) did not affect particle assembly, virus-like particle formation and viral replication in culture cells. However, activation of the ER stress response was significantly reduced in cells infected with rN3D/N6D, correlated with a lower level of apoptosis and reduced production of pro-inflammatory cytokines. Taken together, this study demonstrates that although not essential for replication, glycosylation in the IBV M protein ectodomain plays important roles in activating ER stress, apoptosis and proinflammatory response, and may contribute to the pathogenesis of IBV.
Keywords: Apoptosis; Coronavirus; ER stress; Glycosylation; Particle assembly; Pro-inflammatory response; Viral pathogenesis.
Copyright © 2019. Published by Elsevier Inc.
Figures

References
-
- Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet. Res. 2007;38:281–297. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

