Innate Immune Signaling Organelles Display Natural and Programmable Signaling Flexibility
- PMID: 30853218
- PMCID: PMC6710629
- DOI: 10.1016/j.cell.2019.01.039
Innate Immune Signaling Organelles Display Natural and Programmable Signaling Flexibility
Abstract
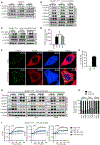
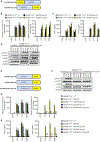
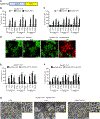
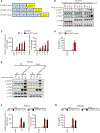
The signaling organelles of the innate immune system consist of oligomeric protein complexes known as supramolecular organizing centers (SMOCs). Examples of SMOCs include myddosomes and inflammasomes, which respectively induce transcription-dependent and -independent inflammatory responses. The common use of oligomeric structures as signaling platforms suggests multifunctionality, but each SMOC has a singular biochemically defined function. Here, we report that the myddosome is a multifunctional organizing center. In addition to promoting inflammatory transcription factor activation, the myddosome drives the rapid induction of glycolysis. We identify the kinase TBK1 as a myddosome component that promotes glycolysis, but not nuclear factor κB (NF-κB) activation. Synthetic immunology approaches further diversified SMOC activities, as we created interferon- or necroptosis-inducing myddosomes, inflammasomes that induce interferon responses instead of pyroptosis, and a SMOC-like nanomachine that induces interferon expression in response to a chemical ligand. These discoveries demonstrate the flexibility of immune signaling organelles, which permits the design of user-defined innate immune responses.
Keywords: MyD88; STING; TBK1; Toll-like Receptors; glycolysis; inflammasome; innate immunity; interferon; myddosome; synthetic biology.
Copyright © 2019 Elsevier Inc. All rights reserved.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

