Unsupervised classification of multi-omics data during cardiac remodeling using deep learning
- PMID: 30853547
- PMCID: PMC6708480
- DOI: 10.1016/j.ymeth.2019.03.004
Unsupervised classification of multi-omics data during cardiac remodeling using deep learning
Abstract
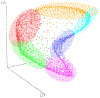
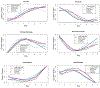
Integration of multi-omics in cardiovascular diseases (CVDs) presents high potentials for translational discoveries. By analyzing abundance levels of heterogeneous molecules over time, we may uncover biological interactions and networks that were previously unidentifiable. However, to effectively perform integrative analysis of temporal multi-omics, computational methods must account for the heterogeneity and complexity in the data. To this end, we performed unsupervised classification of proteins and metabolites in mice during cardiac remodeling using two innovative deep learning (DL) approaches. First, long short-term memory (LSTM)-based variational autoencoder (LSTM-VAE) was trained on time-series numeric data. The low-dimensional embeddings extracted from LSTM-VAE were then used for clustering. Second, deep convolutional embedded clustering (DCEC) was applied on images of temporal trends. Instead of a two-step procedure, DCEC performes a joint optimization for image reconstruction and cluster assignment. Additionally, we performed K-means clustering, partitioning around medoids (PAM), and hierarchical clustering. Pathway enrichment analysis using the Reactome knowledgebase demonstrated that DL methods yielded higher numbers of significant biological pathways than conventional clustering algorithms. In particular, DCEC resulted in the highest number of enriched pathways, suggesting the strength of its unified framework based on visual similarities. Overall, unsupervised DL is shown to be a promising analytical approach for integrative analysis of temporal multi-omics.
Keywords: Cardiovascular; Clustering; Integrative analysis; Multi-omics; Time-series; Unsupervised deep learning.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


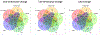
References
-
- Benjamin EJ; Virani SS; Callaway CW; Chamberlain AM; Chang AR; Cheng S; Chiuve SE; Cushman M; Delling FN; Deo R Heart disease and stroke statistics—2018 update: a report from the American Heart Association. Circulation 2018, 137, e67–e492. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

