Uncovering the Mechanisms of Halotolerance in the Extremely Acidophilic Members of the Acidihalobacter Genus Through Comparative Genome Analysis
- PMID: 30853944
- PMCID: PMC6396713
- DOI: 10.3389/fmicb.2019.00155
Uncovering the Mechanisms of Halotolerance in the Extremely Acidophilic Members of the Acidihalobacter Genus Through Comparative Genome Analysis
Abstract
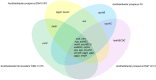
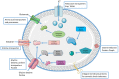
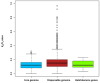
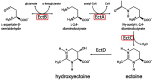
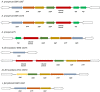
There are few naturally occurring environments where both acid and salinity stress exist together, consequently, there has been little evolutionary pressure for microorganisms to develop systems that enable them to deal with both stresses simultaneously. Members of the genus Acidihalobacter are iron- and sulfur-oxidizing, halotolerant acidophiles that have developed the ability to tolerate acid and saline stress and, therefore, have the potential to bioleach ores with brackish or saline process waters under acidic conditions. The genus consists of four members, A. prosperus DSM 5130T, A. prosperus DSM 14174, A. prosperus F5 and "A. ferrooxidans" DSM 14175. An in depth genome comparison was undertaken in order to provide a more comprehensive description of the mechanisms of halotolerance used by the different members of this genus. Pangenome analysis identified 29, 3 and 9 protein families related to halotolerance in the core, dispensable and unique genomes, respectively. The genes for halotolerance showed Ka/Ks ratios between 0 and 0.2, confirming that they are conserved and stabilized. All the Acidihalobacter genomes contained similar genes for the synthesis and transport of ectoine, which was recently found to be the dominant osmoprotectant in A. prosperus DSM 14174 and A. prosperus DSM 5130T. Similarities also existed in genes encoding low affinity potassium pumps, however, A. prosperus DSM 14174 was also found to contain genes encoding high affinity potassium pumps. Furthermore, only A. prosperus DSM 5130T and "A. ferrooxidans" DSM 14175 contained genes allowing the uptake of taurine as an osmoprotectant. Variations were also seen in genes encoding proteins involved in the synthesis and/or transport of periplasmic glucans, sucrose, proline, and glycine betaine. This suggests that versatility exists in the Acidihalobacter genus in terms of the mechanisms they can use for halotolerance. This information is useful for developing hypotheses for the search for life on exoplanets and moons.
Keywords: Acidihalobacter; acidophile; astrobiology; ectoine; halophile; osmoprotectant.
Figures

References
-
- Alexander B., Leach S., Ingledew W. J. (1987). The relationship between chemiosmotic parameters and sensitivity to anions and organic acids in the acidophile Thiobacillus ferrooxidans. Microbiology 133 1171–1179. 10.1099/00221287-133-5-1171 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

