The comparative genomics and complex population history of Papio baboons
- PMID: 30854422
- PMCID: PMC6401983
- DOI: 10.1126/sciadv.aau6947
The comparative genomics and complex population history of Papio baboons
Abstract
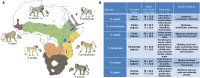
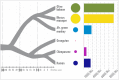
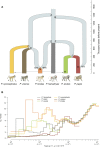
Recent studies suggest that closely related species can accumulate substantial genetic and phenotypic differences despite ongoing gene flow, thus challenging traditional ideas regarding the genetics of speciation. Baboons (genus Papio) are Old World monkeys consisting of six readily distinguishable species. Baboon species hybridize in the wild, and prior data imply a complex history of differentiation and introgression. We produced a reference genome assembly for the olive baboon (Papio anubis) and whole-genome sequence data for all six extant species. We document multiple episodes of admixture and introgression during the radiation of Papio baboons, thus demonstrating their value as a model of complex evolutionary divergence, hybridization, and reticulation. These results help inform our understanding of similar cases, including modern humans, Neanderthals, Denisovans, and other ancient hominins.
Figures

References
-
- Seehausen O., Butlin R. K., Keller I., Wagner C. E., Boughman J. W., Hohenlohe P. A., Peichel C. L., Saetre G.-P., Bank C., Brännström Å., Brelsford A., Clarkson C. S., Eroukhmanoff F., Feder J. L., Fischer M. C., Foote A. D., Franchini P., Jiggins C. D., Jones F. C., Lindholm A. K., Lucek K., Maan M. E., Marques D. A., Martin S. H., Matthews B., Meier J. I., Möst M., Nachman M. W., Nonaka E., Rennison D. J., Schwarzer J., Watson E. T., Westram A. M., Widmer A., Genomics and the origin of species. Nat. Rev. Genet. 15, 176–192 (2014). - PubMed
-
- Wolf J. B. W., Ellegren H., Making sense of genomic islands of differentiation in light of speciation. Nat. Rev. Genet. 18, 87–100 (2017). - PubMed
-
- D. Otte, J. A. Endler, Speciation and Its Consequences (Sinauer Associates, Inc., 1989).
-
- E. Mayr, Systematics and the Origin of Species (Columbia Univ. Press, 1942).
-
- M. L. Arnold, Divergence with Genetic Exchange (Oxford Univ. Press, 2015).

