Accumulation of genetic and epigenetic alterations in normal cells and cancer risk
- PMID: 30854468
- PMCID: PMC6403339
- DOI: 10.1038/s41698-019-0079-0
Accumulation of genetic and epigenetic alterations in normal cells and cancer risk
Abstract
Cancers develop due to the accumulation of genetic and epigenetic alterations. Genetic alterations are induced by aging, mutagenic chemicals, ultraviolet light, and other factors; whereas, epigenetic alterations are mainly by aging and chronic inflammation. The accumulation and patterns of alterations in normal cells reflect our past exposure levels and life history. Most accumulated alterations are considered as passengers, but their accumulation is correlated with cancer drivers. This has been shown for aberrant DNA methylation but has only been speculated for genetic alterations. However, recent technological advancements have enabled measurement of rare point mutations, and studies have shown that their accumulation levels are indeed correlated with cancer risk. When the accumulation levels of aberrant DNA methylation and point mutations are combined, risk prediction becomes even more accurate. When high levels of alterations accumulate, the tissue has a high risk of developing cancer or even multiple cancers and is considered as a "cancerization field", with or without expansion of physiological patches of clonal cells. In this review, we describe the formation of a cancerization field and how we can apply its detection in precision cancer risk diagnosis.
Conflict of interest statement
T.U. is a recipient of funding from Sysmex and Miraca Inc., and has filed a patent for detecting rare point mutations using a small number of sequencing templates.
Figures
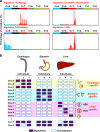
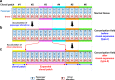
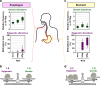
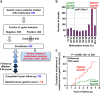
References
-
- Reardon JT, Bessho T, Kung HC, Bolton PH, Sancar A. In vitro repair of oxidative DNA damage by human nucleotide excision repair system: possible explanation for neurodegeneration in xeroderma pigmentosum patients. Proc. Natl. Acad. Sci. USA. 1997;94:9463–9468. doi: 10.1073/pnas.94.17.9463. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

