ITPase deficiency causes a Martsolf-like syndrome with a lethal infantile dilated cardiomyopathy
- PMID: 30856165
- PMCID: PMC6428344
- DOI: 10.1371/journal.pgen.1007605
ITPase deficiency causes a Martsolf-like syndrome with a lethal infantile dilated cardiomyopathy
Abstract
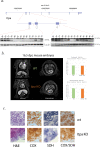
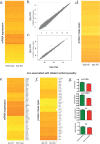
Typical Martsolf syndrome is characterized by congenital cataracts, postnatal microcephaly, developmental delay, hypotonia, short stature and biallelic hypomorphic mutations in either RAB3GAP1 or RAB3GAP2. Genetic analysis of 85 unrelated "mutation negative" probands with Martsolf or Martsolf-like syndromes identified two individuals with different homozygous null mutations in ITPA, the gene encoding inosine triphosphate pyrophosphatase (ITPase). Both probands were from multiplex families with a consistent, lethal and highly distinctive disorder; a Martsolf-like syndrome with infantile-onset dilated cardiomyopathy. Severe ITPase-deficiency has been previously reported with infantile epileptic encephalopathy (MIM 616647). ITPase acts to prevent incorporation of inosine bases (rI/dI) into RNA and DNA. In Itpa-null cells dI was undetectable in genomic DNA. dI could be identified at a low level in mtDNA without detectable mitochondrial genome instability, mtDNA depletion or biochemical dysfunction of the mitochondria. rI accumulation was detectable in proband-derived lymphoblastoid RNA. In Itpa-null mouse embryos rI was detectable in the brain and kidney with the highest level seen in the embryonic heart (rI at 1 in 385 bases). Transcriptome and proteome analysis in mutant cells revealed no major differences with controls. The rate of transcription and the total amount of cellular RNA also appeared normal. rI accumulation in RNA-and by implication rI production-correlates with the severity of organ dysfunction in ITPase deficiency but the basis of the cellulopathy remains cryptic. While we cannot exclude cumulative minor effects, there are no major anomalies in the production, processing, stability and/or translation of mRNA.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Warburg M, Sjo O, Fledelius HC, Pedersen SA. Autosomal recessive microcephaly, microcornea, congenital cataract, mental retardation, optic atrophy, and hypogenitalism. Micro syndrome. Am J Dis Child. 1993;147(12):1309–12. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

