Development of CRISPR/Cas9-mediated gene disruption systems in Giardia lamblia
- PMID: 30856211
- PMCID: PMC6411161
- DOI: 10.1371/journal.pone.0213594
Development of CRISPR/Cas9-mediated gene disruption systems in Giardia lamblia
Abstract
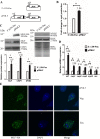
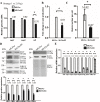
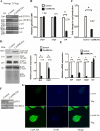
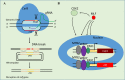
Giardia lamblia becomes dormant by differentiation into a water-resistant cyst that can infect a new host. Synthesis of three cyst wall proteins (CWPs) is the fundamental feature of this differentiation. Myeloid leukemia factor (MLF) proteins are involved in cell differentiation, and tumorigenesis in mammals, but little is known about its role in protozoan parasites. We developed a CRISPR/Cas9 system to understand the role of MLF in Giardia. Due to the tetraploid genome in two nuclei of Giardia, it could be hard to disrupt a gene completely in Giardia. We only generated knockdown but not knockout mutants. We found that knockdown of the mlf gene resulted in a significant decrease of cwp gene expression and cyst formation, suggesting a positive role of MLF in encystation. We further used mlf as a model gene to improve the system. The addition of an inhibitor for NHEJ, Scr7, or combining all cassettes for gRNA and Cas9 expression into one plasmid resulted in improved gene disruption efficiencies and a significant decrease in cwp gene expression. Our results provide insights into a positive role of MLF in inducing Giardia differentiation and a useful tool for studies in Giardia.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
A Novel Multiprotein Bridging Factor 1-Like Protein Induces Cyst Wall Protein Gene Expression and Cyst Differentiation in Giardia lamblia.Int J Mol Sci. 2021 Jan 29;22(3):1370. doi: 10.3390/ijms22031370. Int J Mol Sci. 2021. PMID: 33573049 Free PMC article.
-
A myeloid leukemia factor homolog involved in encystation-induced protein metabolism in Giardia lamblia.Biochim Biophys Acta Gen Subj. 2021 Jun;1865(6):129859. doi: 10.1016/j.bbagen.2021.129859. Epub 2021 Feb 11. Biochim Biophys Acta Gen Subj. 2021. PMID: 33581251
-
A Novel Spo11 Homologue Functions as a Positive Regulator in Cyst Differentiation in Giardia lamblia.Int J Mol Sci. 2021 Nov 2;22(21):11902. doi: 10.3390/ijms222111902. Int J Mol Sci. 2021. PMID: 34769330 Free PMC article.
-
Mechanisms of adaptation in the intestinal parasite Giardia lamblia.Essays Biochem. 2011;51:177-91. doi: 10.1042/bse0510177. Essays Biochem. 2011. PMID: 22023449 Review.
-
Encystation of Giardia lamblia: a model for other parasites.Curr Opin Microbiol. 2007 Dec;10(6):554-9. doi: 10.1016/j.mib.2007.09.011. Epub 2007 Nov 5. Curr Opin Microbiol. 2007. PMID: 17981075 Free PMC article. Review.
Cited by
-
Mlf mediates proteotoxic response via formation of cellular foci for protein folding and degradation in Giardia.PLoS Pathog. 2024 Oct 21;20(10):e1012617. doi: 10.1371/journal.ppat.1012617. eCollection 2024 Oct. PLoS Pathog. 2024. PMID: 39432513 Free PMC article.
-
DNA topoisomerase IIIβ promotes cyst generation by inducing cyst wall protein gene expression in Giardia lamblia.Open Biol. 2020 Feb;10(2):190228. doi: 10.1098/rsob.190228. Epub 2020 Feb 5. Open Biol. 2020. PMID: 32019477 Free PMC article.
-
A myeloid leukemia factor homolog is involved in tolerance to stresses and stress-induced protein metabolism in Giardia lamblia.Biol Direct. 2023 Apr 24;18(1):20. doi: 10.1186/s13062-023-00378-6. Biol Direct. 2023. PMID: 37095576 Free PMC article.
-
Harnessing the power of new genetic tools to illuminate Giardia biology and pathogenesis.Genetics. 2024 Jun 5;227(2):iyae038. doi: 10.1093/genetics/iyae038. Genetics. 2024. PMID: 38626297 Free PMC article. Review.
-
High Cysteine Membrane Proteins (HCMPs) Are Up-Regulated During Giardia-Host Cell Interactions.Front Genet. 2020 Aug 18;11:913. doi: 10.3389/fgene.2020.00913. eCollection 2020. Front Genet. 2020. PMID: 33014015 Free PMC article.
References
-
- Berkman DS, Lescano AG, Gilman RH, Lopez SL, Black MM. Effects of stunting, diarrhoeal disease, and parasitic infection during infancy on cognition in late childhood: a follow-up study. Lancet. 2002;359(9036):564–571. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

