Comparative proteomic analysis provides new insight into differential transmission of two begomoviruses by a whitefly
- PMID: 30857562
- PMCID: PMC6413443
- DOI: 10.1186/s12985-019-1138-4
Comparative proteomic analysis provides new insight into differential transmission of two begomoviruses by a whitefly
Abstract
Background: Viruses in the genus Begomovirus (Family Geminiviridae) include many important economic plant viruses transmitted by whiteflies of the Bemisia tabaci species complex. In general, different begomoviruses may be acquired and transmitted by the same whitefly species with different efficiencies. For example, the species Mediterranean (MED) in this whitefly species complex transmits tomato yellow leaf curl virus (TYLCV) at a higher efficiency than papaya leaf curl China virus (PaLCuCNV). However, the proteomic responses of whitefly to the infection of different begomoviruses remain largely unknown.
Methods: We used iTRAQ-based proteomics coupled with RT-qPCR to investigate and compare responses of the MED whitefly to the infection of TYLCV and PaLCuCNV.
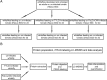
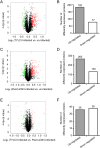
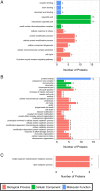
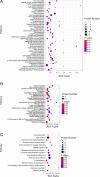
Results: Totally, 259, 395 and 74 differently expressed proteins (DEPs) were identified in the comparisons of TYLCV-infected vs. un-infected, PaLCuCNV-infected vs. un-infected, and TYLCV-infected vs. PaLCuCNV-infected whiteflies, respectively. These proteins appear associated with catabolic process, metabolic process, transport, defense response, cell cycle, and receptor. The comparisons of TYLCV-infected vs. un-infected and PaLCuCNV-infected vs. un-infected shared some similar DEPs, indicating possible involvement of laminin subunit alpha, dystroglycan, integrin alpha-PS2 and cuticle proteins in viral transport as well as the role of putative defense proteins 3 and PITH in anti-viral response. However, 20S proteasome subunits associated with regulation of virus degradation and accumulation were up-regulated in PaLCuCNV-infected but not in TYLCV-infected whiteflies, which may be related to the constraints of PaLCuCNV accumulation in MED.
Conclusions: These findings provide valuable clues for unravelling the roles of some whitefly proteins in begomovirus transmission.
Keywords: Begomovirus; Virus transmission; Whitefly; iTRAQ-based proteomics.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Rosen R, Kanakala S, Kliot A, Pakkianathan BC, Farich BC, Santana-Magal N, Elimelech M, Kontsedalov S, Lebedev G, Cilia M, Ghanim M. Persistent, circulative transmission of begomoviruses by whitefly vector. Curr Opin Virol. 2015;15(1–8). - PubMed
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical

