A complex containing the O-GlcNAc transferase OGT-1 and the ubiquitin ligase EEL-1 regulates GABA neuron function
- PMID: 30858176
- PMCID: PMC6497937
- DOI: 10.1074/jbc.RA119.007406
A complex containing the O-GlcNAc transferase OGT-1 and the ubiquitin ligase EEL-1 regulates GABA neuron function
Abstract
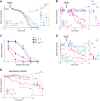
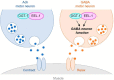
Inhibitory GABAergic transmission is required for proper circuit function in the nervous system. However, our understanding of molecular mechanisms that preferentially influence GABAergic transmission, particularly presynaptic mechanisms, remains limited. We previously reported that the ubiquitin ligase EEL-1 preferentially regulates GABAergic presynaptic transmission. To further explore how EEL-1 functions, here we performed affinity purification proteomics using Caenorhabditis elegans and identified the O-GlcNAc transferase OGT-1 as an EEL-1 binding protein. This observation was intriguing, as we know little about how OGT-1 affects neuron function. Using C. elegans biochemistry, we confirmed that the OGT-1/EEL-1 complex forms in neurons in vivo and showed that the human orthologs, OGT and HUWE1, also bind in cell culture. We observed that, like EEL-1, OGT-1 is expressed in GABAergic motor neurons, localizes to GABAergic presynaptic terminals, and functions cell-autonomously to regulate GABA neuron function. Results with catalytically inactive point mutants indicated that OGT-1 glycosyltransferase activity is dispensable for GABA neuron function. Consistent with OGT-1 and EEL-1 forming a complex, genetic results using automated, behavioral pharmacology assays showed that ogt-1 and eel-1 act in parallel to regulate GABA neuron function. These findings demonstrate that OGT-1 and EEL-1 form a conserved signaling complex and function together to affect GABA neuron function.
Keywords: Caenorhabditis elegans (C. elegans); E3 ubiquitin ligase; EEL-1; GABAergic transmission; HUWE1; O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT); OGT-1; gamma-aminobutyric acid (GABA); motor neuron; neurotransmission; proteomics; synapse.
© 2019 Giles et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

