UVB-induced inactivation of manganese-containing superoxide dismutase promotes mitophagy via ROS-mediated mTORC2 pathway activation
- PMID: 30858178
- PMCID: PMC6497943
- DOI: 10.1074/jbc.RA118.006595
UVB-induced inactivation of manganese-containing superoxide dismutase promotes mitophagy via ROS-mediated mTORC2 pathway activation
Abstract
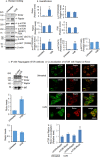
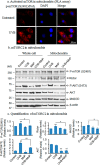
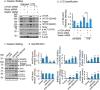
Mitochondria are major sites of energy metabolism that influence numerous cellular events, including immunity and cancer development. Previously, we reported that the mitochondrion-specific antioxidant enzyme, manganese-containing superoxide dismutase (MnSOD), has dual roles in early- and late-carcinogenesis stages. However, how defective MnSOD impacts the chain of events that lead to cell transformation in pathologically normal epidermal cells that have been exposed to carcinogens is unknown. Here, we show that UVB radiation causes nitration and inactivation of MnSOD leading to mitochondrial injury and mitophagy. In keratinocytes, exposure to UVB radiation decreased mitochondrial oxidative phosphorylation, increased glycolysis and the expression of autophagy-related genes, and enhanced AKT Ser/Thr kinase (AKT) phosphorylation and cell growth. Interestingly, UVB initiated a prosurvival mitophagy response by mitochondria-mediated reactive oxygen species (ROS) signaling via the mammalian target of the mTOR complex 2 (mTORC2) pathway. Knockdown of rictor but not raptor abrogated UVB-induced mitophagy responses. Furthermore, fractionation and proximity-ligation assays reveal that ROS-mediated mTOC2 activation in mitochondria is necessary for UVB-induced mitophagy. Importantly, pretreatment with the MnSOD mimic MnTnBuOE-2-PyP5+ (MnP) attenuates mTORC2 activation and suppresses UVB-induced mitophagy. UVB radiation exposure also increased cell growth as assessed by soft-agar colony survival and cell growth assays, and pretreatment with MnP or the known autophagy inhibitor 3-methyladenine abrogated UVB-induced cell growth. These results indicate that MnSOD is a major redox regulator that maintains mitochondrial health and show that UVB-mediated MnSOD inactivation promotes mitophagy and thereby prevents accumulation of damaged mitochondria.
Keywords: MnSOD; autophagy; cancer; mTOR; mTOR complex 2 (mTOR C2); metabolism; mitophagy; oxidative stress; reactive oxygen species (ROS); rictor.
© 2019 Dhar et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




Similar articles
-
A novel MTORC2-AKT-ROS axis triggers mitofission and mitophagy-associated execution of colorectal cancer cells upon drug-induced activation of mutant KRAS.Autophagy. 2024 Jun;20(6):1418-1441. doi: 10.1080/15548627.2024.2307224. Epub 2024 Feb 25. Autophagy. 2024. PMID: 38261660 Free PMC article.
-
DNA polymerase gamma (Polγ) deficiency triggers a selective mTORC2 prosurvival autophagy response via mitochondria-mediated ROS signaling.Oncogene. 2018 Nov;37(48):6225-6242. doi: 10.1038/s41388-018-0404-z. Epub 2018 Jul 23. Oncogene. 2018. PMID: 30038268 Free PMC article.
-
Mitochondrial Perturbations Couple mTORC2 to Autophagy in C. elegans.Cell Rep. 2019 Nov 5;29(6):1399-1409.e5. doi: 10.1016/j.celrep.2019.09.072. Cell Rep. 2019. PMID: 31693882
-
Invited review: manganese superoxide dismutase in disease.Free Radic Res. 2001 Apr;34(4):325-36. doi: 10.1080/10715760100300281. Free Radic Res. 2001. PMID: 11328670 Review.
-
Managing odds in stem cells: insights into the role of mitochondrial antioxidant enzyme MnSOD.Free Radic Res. 2016;50(5):570-84. doi: 10.3109/10715762.2016.1155708. Free Radic Res. 2016. PMID: 26899340 Review.
Cited by
-
NEAT1 Confers Radioresistance to Hepatocellular Carcinoma Cells by Inducing PINK1/Parkin-Mediated Mitophagy.Int J Mol Sci. 2022 Nov 19;23(22):14397. doi: 10.3390/ijms232214397. Int J Mol Sci. 2022. PMID: 36430876 Free PMC article.
-
Inhibitory Effect of Ursolic Acid on Ultraviolet B Radiation-Induced Oxidative Stress and Proinflammatory Response-Mediated Senescence in Human Skin Dermal Fibroblasts.Oxid Med Cell Longev. 2020 Jun 15;2020:1246510. doi: 10.1155/2020/1246510. eCollection 2020. Oxid Med Cell Longev. 2020. PMID: 32617130 Free PMC article.
-
Elimination of damaged mitochondria during UVB-induced senescence is orchestrated by NIX-dependent mitophagy.Aging Cell. 2024 Aug;23(8):e14186. doi: 10.1111/acel.14186. Epub 2024 May 17. Aging Cell. 2024. PMID: 38761001 Free PMC article.
-
Autophagy Contributes to the Quality Control of Leaf Mitochondria.Plant Cell Physiol. 2021 May 11;62(2):229-247. doi: 10.1093/pcp/pcaa162. Plant Cell Physiol. 2021. PMID: 33355344 Free PMC article.
-
Nitric Oxide-Dependent Protein Post-Translational Modifications Impair Mitochondrial Function and Metabolism to Contribute to Neurodegenerative Diseases.Antioxid Redox Signal. 2020 Apr 20;32(12):817-833. doi: 10.1089/ars.2019.7916. Epub 2019 Dec 3. Antioxid Redox Signal. 2020. PMID: 31657228 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

