The ubiquitin interacting motifs of USP37 act on the proximal Ub of a di-Ub chain to enhance catalytic efficiency
- PMID: 30858488
- PMCID: PMC6412040
- DOI: 10.1038/s41598-019-40815-z
The ubiquitin interacting motifs of USP37 act on the proximal Ub of a di-Ub chain to enhance catalytic efficiency
Abstract
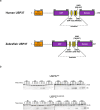
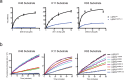
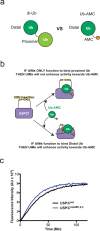
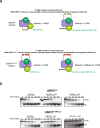
USP37 is a deubiquitinase (DUB) with roles in the regulation of DNA damage repair and the cohesion of sister chromatids during mitosis. USP37 contains a unique insert of three ubiquitin interacting motifs (UIMs) within its catalytic DUB domain. We investigated the role of the three UIMs in the ability of USP37 to cleave di-ubiquitin chains. We found that the third UIM of USP37 recognizes the proximal ubiquitin moiety of K48 di-Ub to potentiate cleavage activity and posit that this mechanism of action may be generalizable to other chain types. In the case of K48-linked ubiquitin chains this potentiation stemmed largely from a dramatic increase in catalytic rate (kcat). We also developed and characterized three ubiquitin variant (UbV) inhibitors that selectively engage distinct binding sites in USP37. In addition to validating the deduced functional roles of the three UIMs in catalysis, the UbVs highlight a novel and effective means to selectively inhibit members of the difficult to drug DUB family.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Ubiquitin-interacting motifs confer full catalytic activity, but not ubiquitin chain substrate specificity, to deubiquitinating enzyme USP37.J Biol Chem. 2014 Jan 24;289(4):2415-23. doi: 10.1074/jbc.M113.528372. Epub 2013 Dec 9. J Biol Chem. 2014. PMID: 24324262 Free PMC article.
-
Panel of Engineered Ubiquitin Variants Targeting the Family of Human Ubiquitin Interacting Motifs.ACS Chem Biol. 2022 Apr 15;17(4):941-956. doi: 10.1021/acschembio.2c00089. Epub 2022 Apr 6. ACS Chem Biol. 2022. PMID: 35385646 Free PMC article.
-
USP37 Deubiquitinates CDC73 in HPT-JT Syndrome.Int J Mol Sci. 2022 Jun 7;23(12):6364. doi: 10.3390/ijms23126364. Int J Mol Sci. 2022. PMID: 35742816 Free PMC article.
-
The Ball and Chain of Polyubiquitin Structures.Trends Biochem Sci. 2016 Apr;41(4):371-385. doi: 10.1016/j.tibs.2016.01.006. Epub 2016 Feb 15. Trends Biochem Sci. 2016. PMID: 26899455 Review.
-
DeSUMOylating enzymes--SENPs.IUBMB Life. 2008 Nov;60(11):734-42. doi: 10.1002/iub.113. IUBMB Life. 2008. PMID: 18666185 Review.
Cited by
-
Ubiquitin Engineering for Interrogating the Ubiquitin-Proteasome System and Novel Therapeutic Strategies.Cells. 2023 Aug 21;12(16):2117. doi: 10.3390/cells12162117. Cells. 2023. PMID: 37626927 Free PMC article. Review.
-
USP37 Promotes Lung Cancer Cell Migration by Stabilizing Snail Protein via Deubiquitination.Front Genet. 2020 Jan 10;10:1324. doi: 10.3389/fgene.2019.01324. eCollection 2019. Front Genet. 2020. PMID: 31998374 Free PMC article.
-
Ubiquitin-specific peptidase 37: an important cog in the oncogenic machinery of cancerous cells.J Exp Clin Cancer Res. 2021 Nov 10;40(1):356. doi: 10.1186/s13046-021-02163-7. J Exp Clin Cancer Res. 2021. PMID: 34758854 Free PMC article. Review.
-
Making Connections: Integrative Signaling Mechanisms Coordinate DNA Break Repair in Chromatin.Front Genet. 2021 Sep 29;12:747734. doi: 10.3389/fgene.2021.747734. eCollection 2021. Front Genet. 2021. PMID: 34659365 Free PMC article. Review.
-
Structural and functional characterization of ubiquitin variant inhibitors for the JAMM-family deubiquitinases STAMBP and STAMBPL1.J Biol Chem. 2021 Oct;297(4):101107. doi: 10.1016/j.jbc.2021.101107. Epub 2021 Aug 21. J Biol Chem. 2021. PMID: 34425109 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

