Vitamin B12 modulates Parkinson's disease LRRK2 kinase activity through allosteric regulation and confers neuroprotection
- PMID: 30858560
- PMCID: PMC6462009
- DOI: 10.1038/s41422-019-0153-8
Vitamin B12 modulates Parkinson's disease LRRK2 kinase activity through allosteric regulation and confers neuroprotection
Abstract
Missense mutations in Leucine-Rich Repeat Kinase 2 (LRRK2) cause the majority of familial and some sporadic forms of Parkinson's disease (PD). The hyperactivity of LRRK2 kinase induced by the pathogenic mutations underlies neurotoxicity, promoting the development of LRRK2 kinase inhibitors as therapeutics. Many potent and specific small-molecule LRRK2 inhibitors have been reported with promise. However, nearly all inhibitors are ATP competitive-some with unwanted side effects and unclear clinical outcome-alternative types of LRRK2 inhibitors are lacking. Herein we identify 5'-deoxyadenosylcobalamin (AdoCbl), a physiological form of the essential micronutrient vitamin B12 as a mixed-type allosteric inhibitor of LRRK2 kinase activity. Multiple assays show that AdoCbl directly binds LRRK2, leading to the alterations of protein conformation and ATP binding in LRRK2. STD-NMR analysis of a LRRK2 homologous kinase reveals the contact sites in AdoCbl that interface with the kinase domain. Furthermore, we provide evidence that AdoCbl modulates LRRK2 activity through disrupting LRRK2 dimerization. Treatment with AdoCbl inhibits LRRK2 kinase activity in cultured cells and brain tissue, and prevents neurotoxicity in cultured primary rodent neurons as well as in transgenic C. elegans and D. melanogaster expressing LRRK2 disease variants. Finally, AdoCbl alleviates deficits in dopamine release sustainability caused by LRRK2 disease variants in mouse models. Our study uncovers vitamin B12 as a novel class of LRRK2 kinase modulator with a distinct mechanism, which can be harnessed to develop new LRRK2-based PD therapeutics in the future.
Conflict of interest statement
The authors declared no competing interest.
Figures
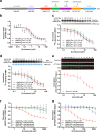
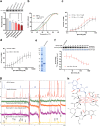
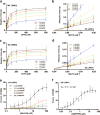
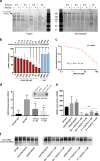
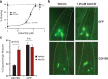
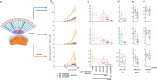

Comment in
-
Linking vitamin B12 and a trembling disorder.Cell Res. 2019 May;29(5):343-344. doi: 10.1038/s41422-019-0166-3. Cell Res. 2019. PMID: 30976075 Free PMC article. No abstract available.
References
-
- Dickson DW, et al. Neuropathology of non-motor features of Parkinson disease. Park. & Relat. Disord. 2009;15(Suppl. 3):S1–S5. - PubMed
-
- Lees AJ, Hardy J, Revesz T. Parkinson’s disease. Lancet. 2009;373:2055–2066. - PubMed
-
- Funayama M, et al. A new locus for Parkinson’s disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann. Neurol. 2002;51:296–301. - PubMed
-
- Paisán-Ruíz C, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. - PubMed
-
- Zimprich A, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

