Statistical power in genome-wide association studies and quantitative trait locus mapping
- PMID: 30858595
- PMCID: PMC6781134
- DOI: 10.1038/s41437-019-0205-3
Statistical power in genome-wide association studies and quantitative trait locus mapping
Abstract
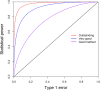
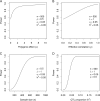
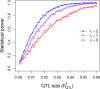
Power calculation prior to a genetic experiment can help investigators choose the optimal sample size to detect a quantitative trait locus (QTL). Without the guidance of power analysis, an experiment may be underpowered or overpowered. Either way will result in wasted resource. QTL mapping and genome-wide association studies (GWAS) are often conducted using a linear mixed model (LMM) with controls of population structure and polygenic background using markers of the whole genome. Power analysis for such a mixed model is often conducted via Monte Carlo simulations. In this study, we derived a non-centrality parameter for the Wald test statistic for association, which allows analytical power analysis. We show that large samples are not necessary to detect a biologically meaningful QTL, say explaining 5% of the phenotypic variance. Several R functions are provided so that users can perform power analysis to determine the minimum sample size required to detect a given QTL with a certain statistical power or calculate the statistical power with given sample size and known values of other population parameters.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Andersen EB. Asymptotic properties of conditional maximum likelihood estimators. J R Stat Soc B. 1970;32:283–301.

