Evaluation of divergent yeast genera for fermentation-associated stresses and identification of a robust sugarcane distillery waste isolate Saccharomyces cerevisiae NGY10 for lignocellulosic ethanol production in SHF and SSF
- PMID: 30858877
- PMCID: PMC6391804
- DOI: 10.1186/s13068-019-1379-x
Evaluation of divergent yeast genera for fermentation-associated stresses and identification of a robust sugarcane distillery waste isolate Saccharomyces cerevisiae NGY10 for lignocellulosic ethanol production in SHF and SSF
Abstract
Background: Lignocellulosic hydrolysates contain a mixture of hexose (C6)/pentose (C5) sugars and pretreatment-generated inhibitors (furans, weak acids and phenolics). Therefore, robust yeast isolates with characteristics of C6/C5 fermentation and tolerance to pretreatment-derived inhibitors are pre-requisite for efficient lignocellulosic material based biorefineries. Moreover, use of thermotolerant yeast isolates will further reduce cooling cost, contamination during fermentation, and required for developing simultaneous saccharification and fermentation (SSF), simultaneous saccharification and co-fermentation (SScF), and consolidated bio-processing (CBP) strategies.
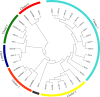
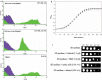
Results: In this study, we evaluated thirty-five yeast isolates (belonging to six genera including Saccharomyces, Kluyveromyces, Candida, Scheffersomyces, Ogatea and Wickerhamomyces) for pretreatment-generated inhibitors {furfural, 5-hydroxymethyl furfural (5-HMF) and acetic acid} and thermotolerant phenotypes along with the fermentation performances at 40 °C. Among them, a sugarcane distillery waste isolate, Saccharomyces cerevisiae NGY10 produced maximum 49.77 ± 0.34 g/l and 46.81 ± 21.98 g/l ethanol with the efficiency of 97.39% and 93.54% at 30 °C and 40 °C, respectively, in 24 h using glucose as a carbon source. Furthermore, isolate NGY10 produced 12.25 ± 0.09 g/l and 7.18 ± 0.14 g/l of ethanol with 92.81% and 91.58% efficiency via SHF, and 30.22 g/l and 25.77 g/l ethanol with 86.43% and 73.29% efficiency via SSF using acid- and alkali-pretreated rice straw as carbon sources, respectively, at 40 °C. In addition, isolate NGY10 also produced 92.31 ± 3.39 g/l (11.7% v/v) and 33.66 ± 1.04 g/l (4.26% v/v) ethanol at 40 °C with the yields of 81.49% and 73.87% in the presence of 30% w/v glucose or 4× concentrated acid-pretreated rice straw hydrolysate, respectively. Moreover, isolate NGY10 displayed furfural- (1.5 g/l), 5-HMF (3.0 g/l), acetic acid- (0.2% v/v) and ethanol-(10.0% v/v) tolerant phenotypes.
Conclusion: A sugarcane distillery waste isolate NGY10 demonstrated high potential for ethanol production, C5 metabolic engineering and developing strategies for SSF, SScF and CBP.
Keywords: Ethanol; Fermentation; Inhibitors; SHF; SSF; Thermo-tolerance.
Figures




Similar articles
-
Bioprospecting thermotolerant ethanologenic yeasts for simultaneous saccharification and fermentation from diverse environments.J Biosci Bioeng. 2017 Mar;123(3):342-346. doi: 10.1016/j.jbiosc.2016.10.007. Epub 2016 Nov 14. J Biosci Bioeng. 2017. PMID: 27856231
-
Bioprospecting thermotolerant yeasts from distillery effluent and molasses for high-temperature ethanol production.J Appl Microbiol. 2022 Feb;132(2):1134-1151. doi: 10.1111/jam.15288. Epub 2021 Sep 15. J Appl Microbiol. 2022. PMID: 34487585
-
Ethanol production from dilute-acid steam exploded lignocellulosic feedstocks using an isolated multistress-tolerant Pichia kudriavzevii strain.Microb Biotechnol. 2017 Nov;10(6):1581-1590. doi: 10.1111/1751-7915.12712. Epub 2017 May 5. Microb Biotechnol. 2017. PMID: 28474425 Free PMC article.
-
Lignocellulosic ethanol: Technology design and its impact on process efficiency.Biotechnol Adv. 2015 Nov 1;33(6 Pt 2):1091-107. doi: 10.1016/j.biotechadv.2014.12.002. Epub 2014 Dec 6. Biotechnol Adv. 2015. PMID: 25485865 Review.
-
Biotechnological strategies to overcome inhibitors in lignocellulose hydrolysates for ethanol production: review.Crit Rev Biotechnol. 2011 Mar;31(1):20-31. doi: 10.3109/07388551003757816. Epub 2010 May 31. Crit Rev Biotechnol. 2011. PMID: 20513164 Review.
Cited by
-
Single yeast cell nanomotions correlate with cellular activity.Sci Adv. 2020 Jun 24;6(26):eaba3139. doi: 10.1126/sciadv.aba3139. eCollection 2020 Jun. Sci Adv. 2020. PMID: 32637604 Free PMC article.
-
Metabolomic Profiling Revealed Diversion of Cytidinediphosphate-Diacylglycerol and Glycerol Pathway towards Denovo Triacylglycerol Synthesis in Rhodosporidium toruloides.J Fungi (Basel). 2021 Nov 13;7(11):967. doi: 10.3390/jof7110967. J Fungi (Basel). 2021. PMID: 34829254 Free PMC article.
-
Xylanolytic and Ethanologenic Potential of Gut Associated Yeasts from Different Species of Termites from India.Mycobiology. 2020 Oct 21;48(6):501-511. doi: 10.1080/12298093.2020.1830742. Mycobiology. 2020. PMID: 33312017 Free PMC article.
-
The Model System Saccharomyces cerevisiae Versus Emerging Non-Model Yeasts for the Production of Biofuels.Life (Basel). 2020 Nov 21;10(11):299. doi: 10.3390/life10110299. Life (Basel). 2020. PMID: 33233378 Free PMC article. Review.
-
Rational engineering of Saccharomyces cerevisiae towards improved tolerance to multiple inhibitors in lignocellulose fermentations.Biotechnol Biofuels. 2021 Aug 28;14(1):173. doi: 10.1186/s13068-021-02021-w. Biotechnol Biofuels. 2021. PMID: 34454598 Free PMC article.
References
-
- Kumar R, Tabatabaei M, Karimi K, Horvath IS. Recent updates on lignocellulosic biomass derived ethanol—a review. Biofuel Res J. 2016;9:347–356. doi: 10.18331/BRJ2016.3.1.4. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

