Tumor suppressor KIF1Bβ regulates mitochondrial apoptosis in collaboration with YME1L1
- PMID: 30859632
- PMCID: PMC6593999
- DOI: 10.1002/mc.22997
Tumor suppressor KIF1Bβ regulates mitochondrial apoptosis in collaboration with YME1L1
Abstract
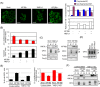
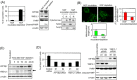
KIF1Bβ, a member of the kinesin superfamily of motor proteins, is a haploinsufficient tumor suppressor mapped to chromosome 1p36.2, which is frequently deleted in neural crest-derived tumors, including neuroblastoma and pheochromocytoma. While KIF1Bβ acts downstream of the nerve growth factor (NGF) pathway to induce apoptosis, further molecular functions of this gene product have largely been unexplored. In this study, we report that KIF1Bβ destabilizes the morphological structure of mitochondria, which is critical for cell survival and apoptosis. We identified YME1L1, a mitochondrial metalloprotease responsible for the cleavage of the mitochondrial GTPase OPA1, as a physical interacting partner of KIF1Bβ. KIF1Bβ interacted with YME1L1 through its death-inducing region, as initiated the protease activity of YME1L1 to cleave the long forms of OPA1, resulting in mitochondrial fragmentation. Overexpression of YME1L1 promoted apoptosis, while knockdown of YME1L1 promoted cell growth. High YME1L1 expression was significantly associated with a better prognosis in neuroblastoma. Furthermore, in NGF-deprived PC12 cells, KIF1Bβ and YME1L1 were upregulated, accompanied by mitochondrial fragmentation and apoptotic cell death. Small interfering RNA-mediated knockdown of either protein alone, however, remarkably inhibited the NGF depletion-induced apoptosis. Our findings indicate that tumor suppressor KIF1Bβ plays an important role in intrinsic mitochondria-mediated apoptosis through the regulation of structural and functional dynamics of mitochondria in collaboration with YME1L1. Dysfunction of the KIF1Bβ/YME1L1/OPA1 mechanism may be involved in malignant biological features of neural crest-derived tumors as well as the initiation and progression of neurodegenerative diseases.
Keywords: KIF1Bβ; YME1L1; mitochondrial fragmentation; neuroblastoma; tumor suppressor.
© 2019 The Authors. Molecular Carcinogenesis Published by Wiley Periodicals, Inc.
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures
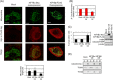
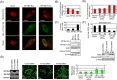
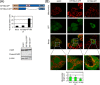

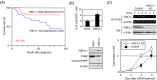
References
-
- Chen H, Chan DC. Emerging functions of mammalian mitochondrial fusion and fission. Hum Mol Genet. 2005;14(Spec No. 2):R283‐R289. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

