Global Outbreaks and Origins of a Chikungunya Virus Variant Carrying Mutations Which May Increase Fitness for Aedes aegypti: Revelations from the 2016 Mandera, Kenya Outbreak
- PMID: 30860010
- PMCID: PMC6493958
- DOI: 10.4269/ajtmh.18-0980
Global Outbreaks and Origins of a Chikungunya Virus Variant Carrying Mutations Which May Increase Fitness for Aedes aegypti: Revelations from the 2016 Mandera, Kenya Outbreak
Abstract
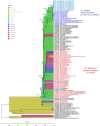
In 2016, a chikungunya virus (CHIKV) outbreak was reported in Mandera, Kenya. This was the first major CHIKV outbreak in the country since the global reemergence of this virus in Kenya in 2004. We collected samples and sequenced viral genomes from this outbreak. All Kenyan genomes contained two mutations, E1:K211E and E2:V264A, recently reported to have an association with increased infectivity, dissemination, and transmission in the Aedes aegypti vector. Phylogeographic inference of temporal and spatial virus relationships showed that this variant emerged within the East, Central, and South African lineage between 2005 and 2008, most probably in India. It was also in India where the first large outbreak caused by this virus appeared, in New Delhi, 2010. More importantly, our results also showed that this variant is no longer contained to India. We found it present in several major outbreaks, including the 2016 outbreaks in Pakistan and Kenya, and the 2017 outbreak in Bangladesh. Thus, this variant may have a capability of driving large CHIKV outbreaks in different regions of the world. Our results point to the importance of continued genomic-based surveillance and prompt urgent vector competence studies to assess the level of vector susceptibility and virus transmission, and the impact this might have on this variant's epidemic potential and global spread.
Figures

Similar articles
-
Emergence of a novel chikungunya virus strain bearing the E1:V80A substitution, out of the Mombasa, Kenya 2017-2018 outbreak.PLoS One. 2020 Nov 6;15(11):e0241754. doi: 10.1371/journal.pone.0241754. eCollection 2020. PLoS One. 2020. PMID: 33156857 Free PMC article.
-
Two novel epistatic mutations (E1:K211E and E2:V264A) in structural proteins of Chikungunya virus enhance fitness in Aedes aegypti.Virology. 2016 Oct;497:59-68. doi: 10.1016/j.virol.2016.06.025. Epub 2016 Jul 15. Virology. 2016. PMID: 27423270
-
Genome sequences of chikungunya virus isolates from an outbreak in southwest Bangkok in 2018.Arch Virol. 2020 Feb;165(2):445-450. doi: 10.1007/s00705-019-04509-1. Epub 2019 Dec 13. Arch Virol. 2020. PMID: 31834526
-
Chikungunya: Its History in Africa and Asia and Its Spread to New Regions in 2013-2014.J Infect Dis. 2016 Dec 15;214(suppl 5):S436-S440. doi: 10.1093/infdis/jiw391. J Infect Dis. 2016. PMID: 27920169 Review.
-
Chikungunya virus infection: molecular biology, clinical characteristics, and epidemiology in Asian countries.J Biomed Sci. 2021 Dec 2;28(1):84. doi: 10.1186/s12929-021-00778-8. J Biomed Sci. 2021. PMID: 34857000 Free PMC article. Review.
Cited by
-
Genetic Characterization of Chikungunya Virus Among Febrile Dengue Fever-Like Patients in Xishuangbanna, Southwestern Part of China.Front Cell Infect Microbiol. 2022 Jun 27;12:914289. doi: 10.3389/fcimb.2022.914289. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35832380 Free PMC article.
-
Complete Coding Sequences of 22 East/Central/South African Genotype Chikungunya Virus Isolates from Thailand (2018 to 2019).Microbiol Resour Announc. 2020 Oct 15;9(42):e00438-20. doi: 10.1128/MRA.00438-20. Microbiol Resour Announc. 2020. PMID: 33060262 Free PMC article.
-
Genome-wide mutational analysis of Chikungunya strains from 2016 to 2017 outbreak of central India: An attempt to elucidate the immunological basis for outbreak.Heliyon. 2022 Nov 4;8(11):e11400. doi: 10.1016/j.heliyon.2022.e11400. eCollection 2022 Nov. Heliyon. 2022. PMID: 36387532 Free PMC article.
-
Large-scale outbreak of Chikungunya virus infection in Thailand, 2018-2019.PLoS One. 2021 Mar 10;16(3):e0247314. doi: 10.1371/journal.pone.0247314. eCollection 2021. PLoS One. 2021. PMID: 33690657 Free PMC article.
-
The global burden of Chikungunya fever among children: A systematic literature review and meta-analysis.PLOS Glob Public Health. 2022 Dec 21;2(12):e0000914. doi: 10.1371/journal.pgph.0000914. eCollection 2022. PLOS Glob Public Health. 2022. PMID: 36962807 Free PMC article.
References
-
- World Health Organization , 2016. Chikungunya-Kenya. Available at: http://www.who.int/csr/don/09-august-2016-chikungunya-kenya/en/. Accessed July 12, 2018.
-
- Sergon K, et al. 2008. Seroprevalence of chikungunya virus (CHIKV) infection on Lamu Island, Kenya, October 2004. Am J Trop Med Hyg 78: 333–337. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

