Comparative Gene-Expression Analysis of Alzheimer's Disease Progression with Aging in Transgenic Mouse Model
- PMID: 30862043
- PMCID: PMC6429175
- DOI: 10.3390/ijms20051219
Comparative Gene-Expression Analysis of Alzheimer's Disease Progression with Aging in Transgenic Mouse Model
Abstract
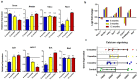
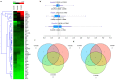
Alzheimer's disease (AD) is a multifactorial neurodegenerative disorder characterized by progressive memory dysfunction and a decline in cognition. One of the biggest challenges to study the pathological process at a molecular level is that there is no simple, cost-effective, and comprehensive gene-expression analysis tool. The present study provides the most detailed (Reverse transcription polymerase chain reaction) RT-PCR-based gene-expression assay, encompassing important genes, based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) disease pathway. This study analyzed age-dependent disease progression by focusing on pathological events such as the processing of the amyloid precursor protein, tau pathology, mitochondrial dysfunction, endoplasmic reticulum stress, disrupted calcium signaling, inflammation, and apoptosis. Messenger RNA was extracted from the cortex and hippocampal region of APP/PS1 transgenic mice. Samples were divided into three age groups, six-, nine-, and 12-month-old transgenic mice, and they were compared with normal C57BL/6J mice of respective age groups. Findings of this study provide the opportunity to design a simple, effective, and accurate clinical analysis tool that can not only provide deeper insight into the disease, but also act as a clinical diagnostic tool for its better diagnosis.
Keywords: Alzheimer’s disease; clinical diagnosis; dementia; gene expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

