Exploiting network topology for large-scale inference of nonlinear reaction models
- PMID: 30862281
- PMCID: PMC6451393
- DOI: 10.1098/rsif.2018.0766
Exploiting network topology for large-scale inference of nonlinear reaction models
Abstract
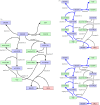
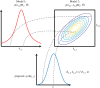
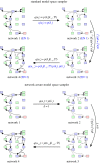
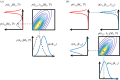
The development of chemical reaction models aids understanding and prediction in areas ranging from biology to electrochemistry and combustion. A systematic approach to building reaction network models uses observational data not only to estimate unknown parameters but also to learn model structure. Bayesian inference provides a natural approach to this data-driven construction of models. Yet traditional Bayesian model inference methodologies that numerically evaluate the evidence for each model are often infeasible for nonlinear reaction network inference, as the number of plausible models can be combinatorially large. Alternative approaches based on model-space sampling can enable large-scale network inference, but their realization presents many challenges. In this paper, we present new computational methods that make large-scale nonlinear network inference tractable. First, we exploit the topology of networks describing potential interactions among chemical species to design improved 'between-model' proposals for reversible-jump Markov chain Monte Carlo. Second, we introduce a sensitivity-based determination of move types which, when combined with network-aware proposals, yields significant additional gains in sampling performance. These algorithms are demonstrated on inference problems drawn from systems biology, with nonlinear differential equation models of species interactions.
Keywords: Bayesian inference; model selection; network inference; reaction network; reversible-jump MCMC.
Conflict of interest statement
We have no competing interests.
Figures

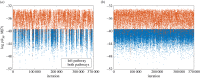
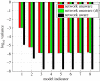
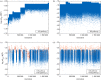

References
-
- Gelman A, Carlin JB, Stern HS, Rudin DB. 2004. Bayesian data analysis, 2nd edn Boca Raton, FL: Chapman and Hall/CRC.
-
- Sachs K, Gifford D, Jaakkola T, Sorger P, Lauffenburger DA. 2002. Bayesian network approach to cell signaling pathway modeling. Sci. STKE 148, pe38. - PubMed
-
- Ellis B, Wong WH. 2008. Learning causal Bayesian network structures from experimental data. J. Am. Stat. Assoc. 103, 778–789. (10.1198/016214508000000193) - DOI
-
- Braman K, Oliver TA, Raman V. 2013. Bayesian analysis of syngas chemistry models. Combust. Theor. Model. 17, 858–887. (10.1080/13647830.2013.811541) - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

