The retroviral accessory proteins S2, Nef, and glycoMA use similar mechanisms for antagonizing the host restriction factor SERINC5
- PMID: 30862674
- PMCID: PMC6497950
- DOI: 10.1074/jbc.RA119.007662
The retroviral accessory proteins S2, Nef, and glycoMA use similar mechanisms for antagonizing the host restriction factor SERINC5
Abstract
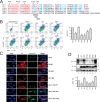
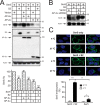
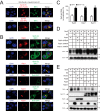
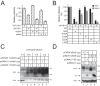
Serine incorporator 5 (SERINC5) is a recently identified restriction factor that blocks virus entry but is antagonized by three unrelated retroviral accessory proteins. The S2 protein from equine infectious anemia virus (EIAV) has been reported to reduce SERINC5 expression at steady-state levels likely via the endocytic pathway; however, the precise mechanism is still unclear. Here, we investigated how EIAV S2 protein down-regulates SERINC5 compared with down-regulation induced by Nef from HIV-1 and glycoMA proteins from murine leukemia virus (MLV). Using bimolecular fluorescence complementation (BiFC) assay and immunoprecipitation (IP), we detected an interaction between S2 and SERINC5. We found that this interaction relies on the S2 myristoylation site, indicating that it may occur on the plasma membrane. S2 internalized SERINC5 via receptor-mediated endocytosis and targeted it to endosomes and lysosomes, resulting in a ubiquitination-dependent decrease in SERINC5 expression at steady-state levels. Both BiFC and IP detected a glycoMA-SERINC5 interaction, but a Nef-SERINC5 interaction was detected only by BiFC. Moreover, S2 and glycoMA down-regulated SERINC5 more effectively than did Nef. We further show that unlike Nef, both S2 and glycoMA effectively down-regulate SERINC2 and also SERINC5 from Xenopus tropicalis (xSERINC5). Moreover, we detected expression of the equine SERINC5 (eSERINC5) protein and observed that its expression is much weaker than expression levels of SERINC5 from other species. Nonetheless, eSERINC5 had a strong antiviral activity that was effectively counteracted by S2. We conclude that HIV-1, EIAV, and MLV share a similar mechanism to antagonize viral restriction by host SERINC5.
Keywords: EIAV S2; HIV Nef; MLV glycoGag; SERINC5; host-pathogen interaction; innate immunity; restriction factor; viral protein; virus; virus entry.
© 2019 Ahmad et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
-
- Rosa A., Chande A., Ziglio S., De Sanctis V., Bertorelli R., Goh S. L., McCauley S. M., Nowosielska A., Antonarakis S. E., Luban J., Santoni F. A., and Pizzato M. (2015) HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526, 212–217 10.1038/nature15399 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

