Phosphorylated tyrosine 93 of hepatitis C virus nonstructural protein 5A is essential for interaction with host c-Src and efficient viral replication
- PMID: 30862675
- PMCID: PMC6509489
- DOI: 10.1074/jbc.RA119.007656
Phosphorylated tyrosine 93 of hepatitis C virus nonstructural protein 5A is essential for interaction with host c-Src and efficient viral replication
Abstract
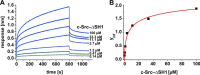
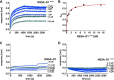
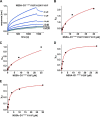
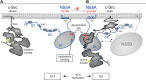
The hepatitis C virus (HCV) nonstructural protein 5A (NS5A) plays a key role in viral replication and virion assembly, and the regulation of the assembly process critically depends on phosphorylation of both serine and threonine residues in NS5A. We previously identified SRC proto-oncogene, nonreceptor tyrosine kinase (c-Src), as an essential host component of the HCV replication complex consisting of NS5A, the RNA-dependent RNA polymerase NS5B, and c-Src. Pulldown assays revealed an interaction between NS5A and the Src homology 2 (SH2) domain of c-Src; however, the precise binding mode remains undefined. In this study, using a variety of biochemical and biophysical techniques, along with molecular dynamics simulations, we demonstrate that the interaction between NS5A and the c-Src SH2 domain strictly depends on an intact phosphotyrosine-binding competent SH2 domain and on tyrosine phosphorylation within NS5A. Detailed analysis of c-Src SH2 domain binding to a panel of phosphorylation-deficient NS5A variants revealed that phosphorylation of Tyr-93 located within domain 1 of NS5A, but not of any other tyrosine residue, is crucial for complex formation. In line with these findings, effective replication of subgenomic HCV replicons as well as production of infectious virus particles in mammalian cell culture models were clearly dependent on the presence of tyrosine at position 93 of NS5A. These findings indicate that phosphorylated Tyr-93 in NS5A plays an important role during viral replication by facilitating NS5A's interaction with the SH2 domain of c-Src.
Keywords: NS5A; Src; Src homology 2 domain (SH2 domain); hepatitis C virus (HCV); liver disease; nonstructural protein 5A; phosphotyrosine; viral replication; virion assembly.
© 2019 Klinker et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
-
- Bode J. G., Brenndörfer E. D., Karthe J., and Häussinger D. (2009) Interplay between host cell and hepatitis C virus in regulating viral replication. Biol. Chem. 390, 1013–1032 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

