Genome-Wide Screening for Enteric Colonization Factors in Carbapenem-Resistant ST258 Klebsiella pneumoniae
- PMID: 30862751
- PMCID: PMC6414703
- DOI: 10.1128/mBio.02663-18
Genome-Wide Screening for Enteric Colonization Factors in Carbapenem-Resistant ST258 Klebsiella pneumoniae
Abstract
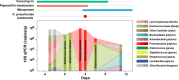
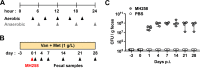
A diverse, antibiotic-naive microbiota prevents highly antibiotic-resistant microbes, including carbapenem-resistant Klebsiella pneumoniae (CR-Kp), from achieving dense colonization of the intestinal lumen. Antibiotic-mediated destruction of the microbiota leads to expansion of CR-Kp in the gut, markedly increasing the risk of bacteremia in vulnerable patients. While preventing dense colonization represents a rational approach to reduce intra- and interpatient dissemination of CR-Kp, little is known about pathogen-associated factors that enable dense growth and persistence in the intestinal lumen. To identify genetic factors essential for dense colonization of the gut by CR-Kp, we constructed a highly saturated transposon mutant library with >150,000 unique mutations in an ST258 strain of CR-Kp and screened for in vitro growth and in vivo intestinal colonization in antibiotic-treated mice. Stochastic and partially reversible fluctuations in the representation of different mutations during dense colonization revealed the dynamic nature of intestinal microbial populations. We identified genes that are crucial for early and late stages of dense gut colonization and confirmed their role by testing isogenic mutants in in vivo competition assays with wild-type CR-Kp Screening of the transposon library also identified mutations that enhanced in vivo CR-Kp growth. These newly identified colonization factors may provide novel therapeutic opportunities to reduce intestinal colonization by CR-KpIMPORTANCEKlebsiella pneumoniae is a common cause of bloodstream infections in immunocompromised and hospitalized patients, and over the last 2 decades, some strains have acquired resistance to nearly all available antibiotics, including broad-spectrum carbapenems. The U.S. Centers for Disease Control and Prevention has listed carbapenem-resistant K. pneumoniae (CR-Kp) as an urgent public health threat. Dense colonization of the intestine by CR-Kp and other antibiotic-resistant bacteria is associated with an increased risk of bacteremia. Reducing the density of gut colonization by CR-Kp is likely to reduce their transmission from patient to patient in health care facilities as well as systemic infections. How CR-Kp expands and persists in the gut lumen, however, is poorly understood. Herein, we generated a highly saturated mutant library in a multidrug-resistant K. pneumoniae strain and identified genetic factors that are associated with dense gut colonization by K. pneumoniae This study sheds light on host colonization by K. pneumoniae and identifies potential colonization factors that contribute to high-density persistence of K. pneumoniae in the intestine.
Keywords: Klebsiella pneumoniae; genome-wide screening; intestinal colonization; multidrug resistance; opportunistic infections.
Copyright © 2019 Jung et al.
Figures



References
-
- Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC. 2001. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161. doi:10.1128/AAC.45.4.1151-1161.2001. - DOI - PMC - PubMed
-
- Snitkin ES, Won S, Pirani A, Lapp Z, Weinstein RA, Lolans K, Hayden MK. 2017. Integrated genomic and interfacility patient-transfer data reveal the transmission pathways of multidrug-resistant Klebsiella pneumoniae in a regional outbreak. Sci Transl Med 9:eaan0093. doi:10.1126/scitranslmed.aan0093. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
