Engulfment, persistence and fate of Bdellovibrio bacteriovorus predators inside human phagocytic cells informs their future therapeutic potential
- PMID: 30862785
- PMCID: PMC6414686
- DOI: 10.1038/s41598-019-40223-3
Engulfment, persistence and fate of Bdellovibrio bacteriovorus predators inside human phagocytic cells informs their future therapeutic potential
Abstract
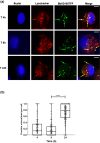
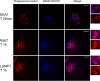
In assessing the potential of predatory bacteria, such as Bdellovibrio bacteriovorus, to become live therapeutic agents against bacterial infections, it is crucial to understand and quantify Bdellovibrio host cell interactions at a molecular level. Here, we quantify the interactions of live B. bacteriovorus with human phagocytic cells, determining the uptake mechanisms, persistence, associated cytokine responses and intracellular trafficking of the non-growing B. bacteriovorus in PMA-differentiated U937 cells. B. bacteriovorus are engulfed by U937 cells and persist for 24 h without affecting host cell viability and can be observed microscopically and recovered and cultured post-uptake. The uptake of predators is passive and depends on the dynamics of the host cell cytoskeleton; the engulfed predators are eventually trafficked through the phagolysosomal pathway of degradation. We have also studied the prevalence of B. bacteriovorus specific antibodies in the general human population. Together, these results quantify a period of viable persistence and the ultimate fate of B. bacteriovorus inside phagocytic cells. They provide new knowledge on predator availability inside hosts, plus potential longevity and therefore potential efficacy as a treatment in humans and open up future fields of work testing if predators can prey on host-engulfed pathogenic bacteria.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- WHO. Antibacterial agents in clinical development: An analysis of the antibacterial clinical development pipeline, including tuberculosis. (2017).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

