Enhanced antibiotic resistance development from fluoroquinolone persisters after a single exposure to antibiotic
- PMID: 30862812
- PMCID: PMC6414640
- DOI: 10.1038/s41467-019-09058-4
Enhanced antibiotic resistance development from fluoroquinolone persisters after a single exposure to antibiotic
Abstract
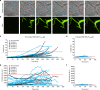
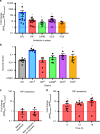
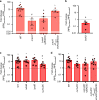
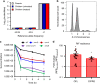
Bacterial persisters are able to tolerate high levels of antibiotics and give rise to new populations. Persister tolerance is generally attributed to minimally active cellular processes that prevent antibiotic-induced damage, which has led to the supposition that persister offspring give rise to antibiotic-resistant mutants at comparable rates to normal cells. Using time-lapse microscopy to monitor Escherichia coli populations following ofloxacin treatment, we find that persisters filament extensively and induce impressive SOS responses before returning to a normal appearance. Further, populations derived from fluoroquinolone persisters contain significantly greater quantities of antibiotic-resistant mutants than those from untreated controls. We confirm that resistance is heritable and that the enhancement requires RecA, SOS induction, an opportunity to recover from treatment, and the involvement of error-prone DNA polymerase V (UmuDC). These findings show that fluoroquinolones damage DNA in persisters and that the ensuing SOS response accelerates the development of antibiotic resistance from these survivors.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

