The X-linked tumor suppressor TSPX downregulates cancer-drivers/oncogenes in prostate cancer in a C-terminal acidic domain dependent manner
- PMID: 30863497
- PMCID: PMC6407674
- DOI: 10.18632/oncotarget.26673
The X-linked tumor suppressor TSPX downregulates cancer-drivers/oncogenes in prostate cancer in a C-terminal acidic domain dependent manner
Abstract
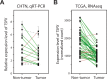
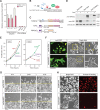
TSPX is a tumor suppressor gene located at Xp11.22, a prostate cancer susceptibility locus. It is ubiquitously expressed in most tissues but frequently downregulated in various cancers, including lung, brain, liver and prostate cancers. The C-terminal acidic domain (CAD) of TSPX is crucial for the tumor suppressor functions, such as inhibition of cyclin B/CDK1 phosphorylation and androgen receptor transactivation. Currently, the exact role of the TSPX CAD in transcriptional regulation of downstream genes is still uncertain. Using different variants of TSPX, we showed that overexpression of either TSPX, that harbors a CAD, or a CAD-truncated variant (TSPX[∆C]) drastically retarded cell proliferation in a prostate cancer cell line LNCaP, but cell death was induced only by overexpression of TSPX. Transcriptome analyses showed that TSPX or TSPX[∆C] overexpression downregulated multiple cancer-drivers/oncogenes, including MYC and MYB, in a CAD-dependent manner and upregulated various tumor suppressors in a CAD-independent manner. Datamining of transcriptomes of prostate cancer specimens in the Cancer Genome Atlas (TCGA) dataset confirmed the negative correlation between the expression level of TSPX and those of MYC and MYB in clinical prostate cancer, thereby supporting the hypothesis that the CAD of TSPX plays an important role in suppression of cancer-drivers/oncogenes in prostatic oncogenesis.
Keywords: TCGA; oncogene; prostate cancer; transcriptome analyses; tumor suppressor.
Conflict of interest statement
CONFLICTS OF INTEREST None.
Figures




References
-
- World Cancer Research Fund International 2018. Available at: http://www.wcrf.org/int/cancer-facts-figures/data-specific-cancers/prost....
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. - PubMed
-
- Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S, Dadaev T, Neal DE, Hamdy FC, et al. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45:385–91. 91e1-2. - PMC - PubMed
-
- Van der Meulen J, Sanghvi V, Mavrakis K, Durinck K, Fang F, Matthijssens F, Rondou P, Rosen M, Pieters T, Vandenberghe P, Delabesse E, Lammens T, De Moerloose B, et al. The H3K27me3 demethylase UTX is a gender-specific tumor suppressor in T-cell acute lymphoblastic leukemia. Blood. 2015;125:13–21. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

