Organelle interplay-peroxisome interactions in health and disease
- PMID: 30864148
- PMCID: PMC7041636
- DOI: 10.1002/jimd.12083
Organelle interplay-peroxisome interactions in health and disease
Abstract
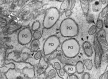
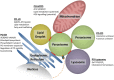
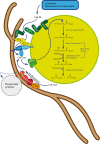
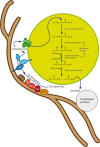
Peroxisomes are multifunctional, dynamic, membrane-bound organelles with important functions in cellular lipid metabolism, rendering them essential for human health and development. Important roles for peroxisomes in signaling and the fine-tuning of cellular processes are emerging, which integrate them in a complex network of interacting cellular compartments. Like many other organelles, peroxisomes communicate through membrane contact sites. For example, peroxisomal growth, positioning, and lipid metabolism involves contacts with the endoplasmic reticulum (ER). Here, we discuss the most recent findings on peroxisome-organelle interactions including peroxisome-ER interplay at membrane contacts sites, and functional interplay with mitochondria, lysosomes, and lipid droplets in mammalian cells. We address tether proteins, metabolic cooperation, and the impact of peroxisome interactions on human health and disease.
Keywords: ACBD5; endoplasmic reticulum; fatty acid beta-oxidation; lipid droplets; lipid metabolism; lysosomes; membrane contact sites; mitochondria; organelle interaction; peroxisomes.
© 2019 The Authors. Journal of Inherited Metabolic Disease published by John Wiley & Sons Ltd on behalf of SSIEM.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Simmen T, Tagaya M. Organelle communication at membrane contact sites (MCS): from curiosity to center stage in cell biology and biomedical research. Adv Exp Med Biol. 2017;997:1‐12. - PubMed
-
- Eisenberg‐Bord M, Shai N, Schuldiner M, Bohnert M. A tether is a tether is a tether: tethering at membrane contact sites. Dev Cell. 2016;39:395‐409. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

