MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization
- PMID: 30864657
- PMCID: PMC6582343
- DOI: 10.1093/nar/gkz173
MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization
Abstract
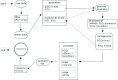
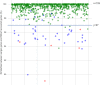
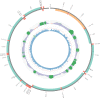
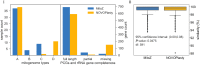
Mitochondrial genome (mitogenome) plays important roles in evolutionary and ecological studies. It becomes routine to utilize multiple genes on mitogenome or the entire mitogenomes to investigate phylogeny and biodiversity of focal groups with the onset of High Throughput Sequencing (HTS) technologies. We developed a mitogenome toolkit MitoZ, consisting of independent modules of de novo assembly, findMitoScaf (find Mitochondrial Scaffolds), annotation and visualization, that can generate mitogenome assembly together with annotation and visualization results from HTS raw reads. We evaluated its performance using a total of 50 samples of which mitogenomes are publicly available. The results showed that MitoZ can recover more full-length mitogenomes with higher accuracy compared to the other available mitogenome assemblers. Overall, MitoZ provides a one-click solution to construct the annotated mitogenome from HTS raw data and will facilitate large scale ecological and evolutionary studies. MitoZ is free open source software distributed under GPLv3 license and available at https://github.com/linzhi2013/MitoZ.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Krehenwinkel H., Kennedy S., Pekár S., Gillespie R.G., Johnston S.. A cost-efficient and simple protocol to enrich prey DNA from extractions of predatory arthropods for large-scale gut content analysis by Illumina sequencing. Methods Ecol. Evol. 2017; 8:126–134.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

