A Listeria monocytogenes ST2 clone lacking chitinase ChiB from an outbreak of non-invasive gastroenteritis
- PMID: 30866756
- PMCID: PMC6455121
- DOI: 10.1080/22221751.2018.1558960
A Listeria monocytogenes ST2 clone lacking chitinase ChiB from an outbreak of non-invasive gastroenteritis
Abstract
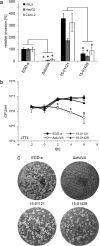
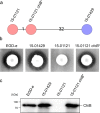
An outbreak with a remarkable Listeria monocytogenes clone causing 163 cases of non-invasive listeriosis occurred in Germany in 2015. Core genome multi locus sequence typing grouped non-invasive outbreak isolates and isolates obtained from related food samples into a single cluster, but clearly separated genetically close isolates obtained from invasive listeriosis cases. A comparative genomic approach identified a premature stop codon in the chiB gene, encoding one of the two L. monocytogenes chitinases, which clustered with disease outcome. Correction of this premature stop codon in one representative gastroenteritis outbreak isolate restored chitinase production, but effects in infection experiments were not found. While the exact role of chitinases in virulence of L. monocytogenes is still not fully understood, our results now clearly show that ChiB-derived activity is not required to establish L. monocytogenes gastroenteritis in humans. This limits a possible role of ChiB in human listeriosis to later steps of the infection.
Keywords: Molecular surveillance; chitin; core genome MLST; invasion; listeriosis.
Figures



References
-
- FAO/WHO Microbiological risk assessment series; no. 5: risk assessment of Listeria monocytogenes in ready-to-eat foods: technical report; 2004. Available from: http://www.fao.org/docrep/pdf/010/y5394e/y5394e.pdf.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
