The hepatitis C virus RNA-dependent RNA polymerase directs incoming nucleotides to its active site through magnesium-dependent dynamics within its F motif
- PMID: 30867194
- PMCID: PMC6514640
- DOI: 10.1074/jbc.RA118.005209
The hepatitis C virus RNA-dependent RNA polymerase directs incoming nucleotides to its active site through magnesium-dependent dynamics within its F motif
Abstract
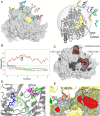
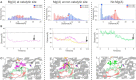
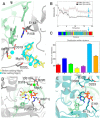
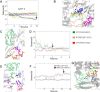
RNA viruses synthesize new genomes in the infected host thanks to dedicated, virally-encoded RNA-dependent RNA polymerases (RdRps). As such, these enzymes are prime targets for antiviral therapy, as has recently been demonstrated for hepatitis C virus (HCV). However, peculiarities in the architecture and dynamics of RdRps raise fundamental questions about access to their active site during RNA polymerization. Here, we used molecular modeling and molecular dynamics simulations, starting from the available crystal structures of HCV NS5B in ternary complex with template-primer duplexes and nucleotides, to address the question of ribonucleotide entry into the active site of viral RdRp. Tracing the possible passage of incoming UTP or GTP through the RdRp-specific entry tunnel, we found two successive checkpoints that regulate nucleotide traffic to the active site. We observed that a magnesium-bound nucleotide first binds next to the tunnel entry, and interactions with the triphosphate moiety orient it such that its base moiety enters first. Dynamics of RdRp motifs F1 + F3 then allow the nucleotide to interrogate the RNA template base prior to nucleotide insertion into the active site. These dynamics are finely regulated by a second magnesium dication, thus coordinating the entry of a magnesium-bound nucleotide with shuttling of the second magnesium necessary for the two-metal ion catalysis. The findings of our work suggest that at least some of these features are general to viral RdRps and provide further details on the original nucleotide selection mechanism operating in RdRps of RNA viruses.
Keywords: RNA virus; RNA-dependent RNA polymerase (RdRp); Single-stranded, positive-sense RNA virus; molecular dynamics; nucleoside/nucleotide transport; protein motif; structural biology; viral polymerase.
© 2019 Ben Ouirane et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

