Integrated Analyses of Phenotype and Quantitative Proteome of CMTM4 Deficient Mice Reveal Its Association with Male Fertility
- PMID: 30867229
- PMCID: PMC6553932
- DOI: 10.1074/mcp.RA119.001416
Integrated Analyses of Phenotype and Quantitative Proteome of CMTM4 Deficient Mice Reveal Its Association with Male Fertility
Abstract
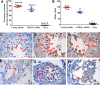
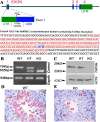
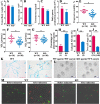
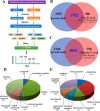
The chemokine-like factor (CKLF)-like MARVEL transmembrane domain-containing family (CMTM) is a gene family that has been implicated in male reproduction. CMTM4 is an evolutionarily conserved member that is highly expressed in the testis. However, its function in male fertility remains unknown. Here, we demonstrate that CMTM4 is associated with spermatogenesis and sperm quality. Using Western blotting and immunohistochemical analyses, we found CMTM4 expression to be decreased in poor-quality human spermatozoa, old human testes, and testicular biopsies with nonobstructive azoospermia. Using CRISPR-Cas9 technology, we knocked out the Cmtm4 gene in mice. These Cmtm4 knockout (KO) mice showed reduced testicular daily sperm production, lower epididymal sperm motility and increased proportion of abnormally backward-curved sperm heads and bent sperm midpieces. These mice also had an evident sub-fertile phenotype, characterized by low pregnancy rates on prolonged breeding with wild type female mice, reduced in vitro fertilization efficiency and a reduced percentage of acrosome reactions. We then performed quantitative proteomic analysis of the testes, where we identified 139 proteins to be downregulated in Cmtm4-KO mice, 100 (71.9%) of which were related to sperm motility and acrosome reaction. The same proteomic analysis was performed on sperm, where we identified 3588 proteins with 409 being differentially regulated in Cmtm4-KO mice. Our enrichment analysis showed that upregulated proteins were enriched with nucleosomal DNA binding functions and the downregulated proteins were enriched with actin binding functions. These findings elucidate the roles of CMTM4 in male fertility and demonstrates its potential as a promising molecular candidate for sperm quality assessment and the diagnosis or treatment of male infertility.
Keywords: Absolute quantification; CMTM4; Cas9; Cytokines*; Knockouts*; Male fertility; Mouse models; Spermatogenesis; Testis; iTRAQ.
© 2019 Liu et al.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Co-expression of sperm membrane proteins CMTM2A and CMTM2B is essential for ADAM3 localization and male fertility in mice.J Cell Sci. 2018 Oct 8;131(19):jcs221481. doi: 10.1242/jcs.221481. J Cell Sci. 2018. PMID: 30209135 Free PMC article.
-
Testis-specific serine kinase 3 is required for sperm morphogenesis and male fertility.Andrology. 2023 Jul;11(5):826-839. doi: 10.1111/andr.13314. Epub 2022 Nov 16. Andrology. 2023. PMID: 36306217 Free PMC article.
-
CMTM4 is frequently downregulated and functions as a tumour suppressor in clear cell renal cell carcinoma.J Exp Clin Cancer Res. 2015 Oct 16;34:122. doi: 10.1186/s13046-015-0236-4. J Exp Clin Cancer Res. 2015. PMID: 26474560 Free PMC article.
-
CMTM6 and CMTM4 as two novel regulators of PD-L1 modulate the tumor microenvironment.Front Immunol. 2022 Jul 25;13:971428. doi: 10.3389/fimmu.2022.971428. eCollection 2022. Front Immunol. 2022. PMID: 35958549 Free PMC article. Review.
-
Sperm Proteome: What Is on the Horizon?Reprod Sci. 2015 Jun;22(6):638-53. doi: 10.1177/1933719114558918. Epub 2014 Nov 5. Reprod Sci. 2015. PMID: 25376881 Review.
Cited by
-
Towards a kingdom of reproductive life - the core sperm proteome.Reproduction. 2025 May 10;169(6):e250105. doi: 10.1530/REP-25-0105. Print 2025 Jun 1. Reproduction. 2025. PMID: 40298292 Free PMC article.
-
Whole-Genome Profile of Greek Patients with Teratozοοspermia: Identification of Candidate Variants and Genes.Genes (Basel). 2022 Sep 8;13(9):1606. doi: 10.3390/genes13091606. Genes (Basel). 2022. PMID: 36140773 Free PMC article.
-
Application of CRISPR/Cas Technology in Spermatogenesis Research and Male Infertility Treatment.Genes (Basel). 2022 Jun 1;13(6):1000. doi: 10.3390/genes13061000. Genes (Basel). 2022. PMID: 35741761 Free PMC article. Review.
-
Overexpressed CMTM6 Improves Prognosis and Associated With Immune Infiltrates of Ovarian Cancer.Front Mol Biosci. 2022 Jan 31;9:769032. doi: 10.3389/fmolb.2022.769032. eCollection 2022. Front Mol Biosci. 2022. PMID: 35174213 Free PMC article.
-
In vivo study revealed pro-tumorigenic effect of CMTM3 in hepatocellular carcinoma involving the regulation of peroxisome proliferator-activated receptor gamma (PPARγ).Cell Oncol (Dordr). 2023 Feb;46(1):49-64. doi: 10.1007/s13402-022-00733-1. Epub 2022 Oct 26. Cell Oncol (Dordr). 2023. PMID: 36284038
References
-
- Han W., Lou Y., Tang J., Zhang Y., Chen Y., Li Y., Gu W., Huang J., Gui L., Tang Y., Li F., Song Q., Di C., Wang L., Shi Q., Sun R., Xia D., Rui M., Tang J., and Ma D. (2001) Molecular cloning and characterization of chemokine-like factor 1 (CKLF1), a novel human cytokine with unique structure and potential chemotactic activity. Biochem. J. 357, 127–135 - PMC - PubMed
-
- Han W., Ding P., Xu M., Wang L., Rui M., Shi S., Liu Y., Zheng Y., Chen Y., Yang T., and Ma D. (2003) Identification of eight genes encoding chemokine-like factor superfamily members 1–8 (CKLFSF1–8) by in silico cloning and experimental validation. Genomics 81, 609–617 - PubMed
-
- Wang Y., Li J., Cui Y., Li T., Ng K. M., Geng H., Li H., Shu X. S., Li H., Liu W., Luo B., Zhang Q., Mok T. S., Zheng W., Qiu X., Srivastava G., Yu J., Sung J. J., Chan A. T., Ma D., Tao Q., and Han W. (2009) CMTM3, located at the critical tumor suppressor locus 16q22.1, is silenced by CpG methylation in carcinomas and inhibits tumor cell growth through inducing apoptosis. Cancer Res. 69, 5194–5201 - PubMed
-
- Shao L., Cui Y., Li H., Liu Y., Zhao H., Wang Y., Zhang Y., Ng K. M., Han W., Ma D., and Tao Q. (2007) CMTM5 exhibits tumor suppressor activities and is frequently silenced by methylation in carcinoma cell lines. Clin. Cancer Res. 13, 5756–5762 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

