The Infectious Bronchitis Coronavirus Envelope Protein Alters Golgi pH To Protect the Spike Protein and Promote the Release of Infectious Virus
- PMID: 30867314
- PMCID: PMC6532078
- DOI: 10.1128/JVI.00015-19
The Infectious Bronchitis Coronavirus Envelope Protein Alters Golgi pH To Protect the Spike Protein and Promote the Release of Infectious Virus
Abstract
Coronaviruses (CoVs) assemble by budding into the lumen of the early Golgi complex prior to exocytosis. The small CoV envelope (E) protein plays roles in assembly, virion release, and pathogenesis. CoV E has a single hydrophobic domain (HD), is targeted to Golgi membranes, and has cation channel activity in vitro The E protein from avian infectious bronchitis virus (IBV) has dramatic effects on the secretory system, which require residues in the HD. Mutation of the HD of IBV E in a recombinant virus background results in impaired growth kinetics, impaired release of infectious virions, accumulation of IBV spike (S) protein on the plasma membrane compared to wild-type (WT) IBV-infected cells, and aberrant cleavage of IBV S on virions. We previously reported the formation of two distinct oligomeric pools of IBV E in transfected and infected cells. Disruption of the secretory pathway by IBV E correlates with a form that is likely monomeric, suggesting that the effects on the secretory pathway are independent of E ion channel activity. Here, we present evidence suggesting that the monomeric form of IBV E correlates with an increased Golgi luminal pH. Infection with IBV or expression of IBV E induces neutralization of Golgi pH, promoting a model in which IBV E alters the secretory pathway through interaction with host cell factors, protecting IBV S from premature cleavage and leading to the efficient release of infectious virus from the cells. This is the first demonstration of a coronavirus-induced alteration in the microenvironment of the secretory pathway.IMPORTANCE Coronaviruses are important human pathogens with significant zoonotic potential. Progress has been made toward identifying potential vaccine candidates for highly pathogenic human CoVs, including the use of attenuated viruses that lack the CoV E protein or express E mutants. However, no approved vaccines or antiviral therapeutics exist. Understanding the role of the CoV E protein in virus assembly and release is thus an important prerequisite for potential vaccines as well as in identifying novel antiviral therapeutics.
Keywords: E protein; Golgi; coronavirus; oligomers; pH; viroporin.
Copyright © 2019 American Society for Microbiology.
Figures
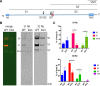
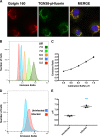
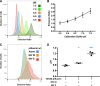

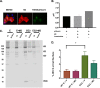
Similar articles
-
A Coronavirus E Protein Is Present in Two Distinct Pools with Different Effects on Assembly and the Secretory Pathway.J Virol. 2015 Sep;89(18):9313-23. doi: 10.1128/JVI.01237-15. Epub 2015 Jul 1. J Virol. 2015. PMID: 26136577 Free PMC article.
-
The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus.J Virol. 2011 Jan;85(2):675-85. doi: 10.1128/JVI.01570-10. Epub 2010 Nov 3. J Virol. 2011. PMID: 21047962 Free PMC article.
-
A single polar residue and distinct membrane topologies impact the function of the infectious bronchitis coronavirus E protein.PLoS Pathog. 2012;8(5):e1002674. doi: 10.1371/journal.ppat.1002674. Epub 2012 May 3. PLoS Pathog. 2012. PMID: 22570613 Free PMC article.
-
Severe acute respiratory syndrome vaccine development: experiences of vaccination against avian infectious bronchitis coronavirus.Avian Pathol. 2003 Dec;32(6):567-82. doi: 10.1080/03079450310001621198. Avian Pathol. 2003. PMID: 14676007 Free PMC article. Review.
-
Incorporation of spike and membrane glycoproteins into coronavirus virions.Viruses. 2015 Apr 3;7(4):1700-25. doi: 10.3390/v7041700. Viruses. 2015. PMID: 25855243 Free PMC article. Review.
Cited by
-
A review on structural, non-structural, and accessory proteins of SARS-CoV-2: Highlighting drug target sites.Immunobiology. 2023 Jan;228(1):152302. doi: 10.1016/j.imbio.2022.152302. Epub 2022 Nov 15. Immunobiology. 2023. PMID: 36434912 Free PMC article. Review.
-
Assembly and Entry of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV2): Evaluation Using Virus-Like Particles.Cells. 2021 Apr 9;10(4):853. doi: 10.3390/cells10040853. Cells. 2021. PMID: 33918600 Free PMC article.
-
Potential Antiviral Strategy Exploiting Dependence of SARS-CoV-2 Replication on Lysosome-Based Pathway.Int J Mol Sci. 2022 May 31;23(11):6188. doi: 10.3390/ijms23116188. Int J Mol Sci. 2022. PMID: 35682877 Free PMC article. Review.
-
The envelope proteins from SARS-CoV-2 and SARS-CoV potently reduce the infectivity of human immunodeficiency virus type 1 (HIV-1).Retrovirology. 2022 Nov 19;19(1):25. doi: 10.1186/s12977-022-00611-6. Retrovirology. 2022. PMID: 36403071 Free PMC article.
-
In Silico Evaluation of Hexamethylene Amiloride Derivatives as Potential Luminal Inhibitors of SARS-CoV-2 E Protein.Int J Mol Sci. 2022 Sep 13;23(18):10647. doi: 10.3390/ijms231810647. Int J Mol Sci. 2022. PMID: 36142556 Free PMC article.
References
-
- Hogue BG, Machamer CE. 2008. Coronavirus structural proteins and virus assembly, p 179–200. In Perlman S, Gallagher T, Snijder EJ(ed), Nidoviruses. ASM Press, Washington, DC.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

