Duodenal bacterial proteolytic activity determines sensitivity to dietary antigen through protease-activated receptor-2
- PMID: 30867416
- PMCID: PMC6416356
- DOI: 10.1038/s41467-019-09037-9
Duodenal bacterial proteolytic activity determines sensitivity to dietary antigen through protease-activated receptor-2
Abstract
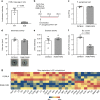
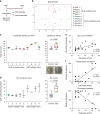
Microbe-host interactions are generally homeostatic, but when dysfunctional, they can incite food sensitivities and chronic diseases. Celiac disease (CeD) is a food sensitivity characterized by a breakdown of oral tolerance to gluten proteins in genetically predisposed individuals, although the underlying mechanisms are incompletely understood. Here we show that duodenal biopsies from patients with active CeD have increased proteolytic activity against gluten substrates that correlates with increased Proteobacteria abundance, including Pseudomonas. Using Pseudomonas aeruginosa producing elastase as a model, we show gluten-independent, PAR-2 mediated upregulation of inflammatory pathways in C57BL/6 mice without villus blunting. In mice expressing CeD risk genes, P. aeruginosa elastase synergizes with gluten to induce more severe inflammation that is associated with moderate villus blunting. These results demonstrate that proteases expressed by opportunistic pathogens impact host immune responses that are relevant to the development of food sensitivities, independently of the trigger antigen.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Bustos D, et al. Colonic proteinases: increased activity in patients with ulcerative colitis. Medicina. 1998;58:262–264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

