Transcriptional profiling of human microglia reveals grey-white matter heterogeneity and multiple sclerosis-associated changes
- PMID: 30867424
- PMCID: PMC6416318
- DOI: 10.1038/s41467-019-08976-7
Transcriptional profiling of human microglia reveals grey-white matter heterogeneity and multiple sclerosis-associated changes
Abstract
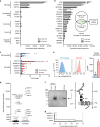
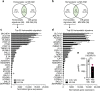
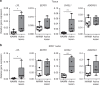
Here we report the transcriptional profile of human microglia, isolated from normal-appearing grey matter (GM) and white matter (WM) of multiple sclerosis (MS) and non-neurological control donors, to find possible early changes related to MS pathology. Microglia show a clear region-specific profile, indicated by higher expression of type-I interferon genes in GM and higher expression of NF-κB pathway genes in WM. Transcriptional changes in MS microglia also differ between GM and WM. MS WM microglia show increased lipid metabolism gene expression, which relates to MS pathology since active MS lesion-derived microglial nuclei show similar altered gene expression. Microglia from MS GM show increased expression of genes associated with glycolysis and iron homeostasis, possibly reflecting microglia reacting to iron depositions. Except for ADGRG1/GPR56, expression of homeostatic genes, such as P2RY12 and TMEM119, is unaltered in normal-appearing MS tissue, demonstrating overall preservation of microglia homeostatic functions in the initiation phase of MS.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Stadelman C, Wegner C, Bruck W. Inflammation, demyelination, and degeneration- recent insights from MS pathology. Biochim. Biophys. Acta. 2011;1812:275–282. - PubMed
-
- Mahad DH, Trapp BD, Lassmann H. Pathological mechanisms in progressive multiple sclerosis. Lancet Neurol. 2015;14:183–193. - PubMed
-
- Peterson JW, Bö L, Mörk S, Chang A, Trapp BD. Transected neurites, apoptotic neurons, and reduced inflammation in cortical multiple sclerosis lesions. Ann. Neurol. 2001;50:389–400. - PubMed
-
- Bø L, Vedeler CA, Nyland H, Trapp BD, Mørk SJ. Intracortical multiple sclerosis lesions are not associated with increased lymphocyte infiltration. Mult. Scler. 2003;9:323–331. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

