A Hox-TALE regulatory circuit for neural crest patterning is conserved across vertebrates
- PMID: 30867425
- PMCID: PMC6416258
- DOI: 10.1038/s41467-019-09197-8
A Hox-TALE regulatory circuit for neural crest patterning is conserved across vertebrates
Abstract
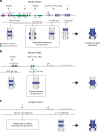
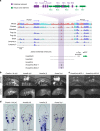
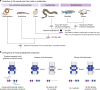
In jawed vertebrates (gnathostomes), Hox genes play an important role in patterning head and jaw formation, but mechanisms coupling Hox genes to neural crest (NC) are unknown. Here we use cross-species regulatory comparisons between gnathostomes and lamprey, a jawless extant vertebrate, to investigate conserved ancestral mechanisms regulating Hox2 genes in NC. Gnathostome Hoxa2 and Hoxb2 NC enhancers mediate equivalent NC expression in lamprey and gnathostomes, revealing ancient conservation of Hox upstream regulatory components in NC. In characterizing a lamprey hoxα2 NC/hindbrain enhancer, we identify essential Meis, Pbx, and Hox binding sites that are functionally conserved within Hoxa2/Hoxb2 NC enhancers. This suggests that the lamprey hoxα2 enhancer retains ancestral activity and that Hoxa2/Hoxb2 NC enhancers are ancient paralogues, which diverged in hindbrain and NC activities. This identifies an ancestral mechanism for Hox2 NC regulation involving a Hox-TALE regulatory circuit, potentiated by inputs from Meis and Pbx proteins and Hox auto-/cross-regulatory interactions.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Le Douarin, N. & Kalcheim, C. The Neural Crest 2nd edn (Cambridge University Press, Cambridge, 1999).
-
- Parker, H. J., Pushel, I. & Krumlauf, R. Coupling the roles of Hox genes to regulatory networks patterning cranial neural crest. Dev. Biol.10.1016/j.ydbio.2018.1003.1016 (2018). - PubMed
-
- Gavalas A, Trainor P, Ariza-McNaughton L, Krumlauf R. Synergy between Hoxa1 and Hoxb1: the relationship between arch patterning and the generation of cranial neural crest. Development. 2001;128:3017–3027. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

