Network-based prediction of drug combinations
- PMID: 30867426
- PMCID: PMC6416394
- DOI: 10.1038/s41467-019-09186-x
Network-based prediction of drug combinations
Erratum in
-
Publisher Correction: Network-based prediction of drug combinations.Nat Commun. 2019 Apr 15;10(1):1806. doi: 10.1038/s41467-019-09692-y. Nat Commun. 2019. PMID: 30988295 Free PMC article.
Abstract
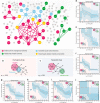
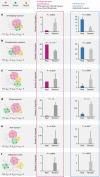
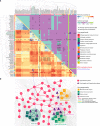
Drug combinations, offering increased therapeutic efficacy and reduced toxicity, play an important role in treating multiple complex diseases. Yet, our ability to identify and validate effective combinations is limited by a combinatorial explosion, driven by both the large number of drug pairs as well as dosage combinations. Here we propose a network-based methodology to identify clinically efficacious drug combinations for specific diseases. By quantifying the network-based relationship between drug targets and disease proteins in the human protein-protein interactome, we show the existence of six distinct classes of drug-drug-disease combinations. Relying on approved drug combinations for hypertension and cancer, we find that only one of the six classes correlates with therapeutic effects: if the targets of the drugs both hit disease module, but target separate neighborhoods. This finding allows us to identify and validate antihypertensive combinations, offering a generic, powerful network methodology to identify efficacious combination therapies in drug development.
Conflict of interest statement
A.-L.B. is a co-founder of Scipher, a startup that uses network concepts to explore human disease. The other authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

