A panel of colorimetric assays to measure enzymatic activity in the base excision DNA repair pathway
- PMID: 30869144
- PMCID: PMC6582407
- DOI: 10.1093/nar/gkz171
A panel of colorimetric assays to measure enzymatic activity in the base excision DNA repair pathway
Abstract
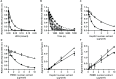
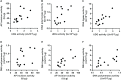
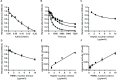
DNA repair is essential for the maintenance of genomic integrity, and evidence suggest that inter-individual variation in DNA repair efficiency may contribute to disease risk. However, robust assays suitable for quantitative determination of DNA repair capacity in large cohort and clinical trials are needed to evaluate these apparent associations fully. We describe here a set of microplate-based oligonucleotide assays for high-throughput, non-radioactive and quantitative determination of repair enzyme activity at individual steps and over multiple steps of the DNA base excision repair pathway. The assays are highly sensitive: using HepG2 nuclear extract, enzyme activities were quantifiable at concentrations of 0.0002 to 0.181 μg per reaction, depending on the enzyme being measured. Assay coefficients of variation are comparable with other microplate-based assays. The assay format requires no specialist equipment and has the potential to be extended for analysis of a wide range of DNA repair enzyme activities. As such, these assays hold considerable promise for gaining new mechanistic insights into how DNA repair is related to individual genetics, disease status or progression and other environmental factors and investigating whether DNA repair activities can be used a biomarker of disease risk.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
Fluorogenic DNA ligase and base excision repair enzyme assays using substrates labeled with single fluorophores.Anal Biochem. 2015 May 15;477:69-77. doi: 10.1016/j.ab.2015.02.022. Epub 2015 Feb 26. Anal Biochem. 2015. PMID: 25728944
-
Analysis of Base Excision and Single-Strand Break Repair Activities in Trypanosomatid Extracts.Methods Mol Biol. 2020;2116:353-364. doi: 10.1007/978-1-0716-0294-2_22. Methods Mol Biol. 2020. PMID: 32221931
-
Label-free colorimetric assay for base excision repair enzyme activity based on nicking enzyme assisted signal amplification.Biosens Bioelectron. 2014 Apr 15;54:598-602. doi: 10.1016/j.bios.2013.11.062. Epub 2013 Nov 27. Biosens Bioelectron. 2014. PMID: 24333571
-
Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage.Toxicology. 2003 Nov 15;193(1-2):43-65. doi: 10.1016/s0300-483x(03)00289-0. Toxicology. 2003. PMID: 14599767 Review.
-
DNA damage excision repair in microplate wells with chemiluminescence detection: development and perspectives.Biochimie. 1999 Jan-Feb;81(1-2):53-8. doi: 10.1016/s0300-9084(99)80038-8. Biochimie. 1999. PMID: 10214910 Review.
Cited by
-
High-throughput analysis of DNA repair in microplates towards identification of inhibitors.Genes Environ. 2020 Mar 9;42:11. doi: 10.1186/s41021-020-00153-3. eCollection 2020. Genes Environ. 2020. PMID: 32165992 Free PMC article.
-
CometChip analysis of human primary lymphocytes enables quantification of inter-individual differences in the kinetics of repair of oxidative DNA damage.Free Radic Biol Med. 2021 Oct;174:89-99. doi: 10.1016/j.freeradbiomed.2021.07.033. Epub 2021 Jul 26. Free Radic Biol Med. 2021. PMID: 34324980 Free PMC article.
-
Horizontal acquisition of a DNA ligase improves DNA damage tolerance in eukaryotes.Nat Commun. 2023 Nov 22;14(1):7638. doi: 10.1038/s41467-023-43075-8. Nat Commun. 2023. PMID: 37993452 Free PMC article.
-
Base-Resolution Analysis of Deoxyuridine at the Genome Scale Based on the Artificial Incorporation Modified Nucleobase.ACS Cent Sci. 2021 Jun 23;7(6):973-979. doi: 10.1021/acscentsci.0c01504. Epub 2021 Apr 28. ACS Cent Sci. 2021. PMID: 34235258 Free PMC article.
-
Colon epithelial cell TGFβ signaling modulates the expression of tight junction proteins and barrier function in mice.Am J Physiol Gastrointest Liver Physiol. 2021 Jun 1;320(6):G936-G957. doi: 10.1152/ajpgi.00053.2021. Epub 2021 Mar 24. Am J Physiol Gastrointest Liver Physiol. 2021. PMID: 33759564 Free PMC article.
References
-
- Jeggo P.A., Pearl L.H., Carr A.M.. DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer. 2016; 16:35–42. - PubMed
-
- D’Errico M., Parlanti E., Pascucci B., Fortini P., Baccarini S., Simonelli V., Dogliotti E.. Single nucleotide polymorphisms in DNA glycosylases: from function to disease. Free Radic. Biol. Med. 2016; 107:278–291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

