A checkpoint-oriented cell cycle simulation model
- PMID: 30870080
- PMCID: PMC6527278
- DOI: 10.1080/15384101.2019.1591125
A checkpoint-oriented cell cycle simulation model
Abstract
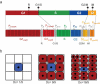
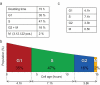
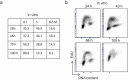
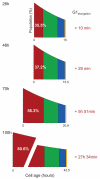
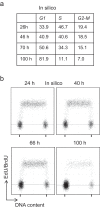
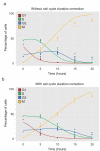
Modeling and in silico simulations are of major conceptual and applicative interest in studying the cell cycle and proliferation in eukaryotic cells. In this paper, we present a cell cycle checkpoint-oriented simulator that uses agent-based simulation modeling to reproduce the dynamics of a cancer cell population in exponential growth. Our in silico simulations were successfully validated by experimental in vitro supporting data obtained with HCT116 colon cancer cells. We demonstrated that this model can simulate cell confluence and the associated elongation of the G1 phase. Using nocodazole to synchronize cancer cells at mitosis, we confirmed the model predictivity and provided evidence of an additional and unexpected effect of nocodazole on the overall cell cycle progression. We anticipate that this cell cycle simulator will be a potential source of new insights and research perspectives.
Keywords: Cell cycle; agent-based modeling; in silico simulation; synchronization.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
