Molecular characterization of Bathymodiolus mussels and gill symbionts associated with chemosynthetic habitats from the U.S. Atlantic margin
- PMID: 30870419
- PMCID: PMC6417655
- DOI: 10.1371/journal.pone.0211616
Molecular characterization of Bathymodiolus mussels and gill symbionts associated with chemosynthetic habitats from the U.S. Atlantic margin
Abstract
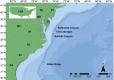
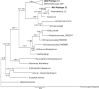
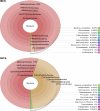
Mussels of the genus Bathymodiolus are among the most widespread colonizers of hydrothermal vent and cold seep environments, sustained by endosymbiosis with chemosynthetic bacteria. Presumed species of Bathymodiolus are abundant at newly discovered cold seeps on the Mid-Atlantic continental slope, however morphological taxonomy is challenging, and their phylogenetic affinities remain unestablished. Here we used mitochondrial sequence to classify species found at three seep sites (Baltimore Canyon seep (BCS; ~400m); Norfolk Canyon seep (NCS; ~1520m); and Chincoteague Island seep (CTS; ~1000m)). Mitochondrial COI (N = 162) and ND4 (N = 39) data suggest that Bathymodiolus childressi predominates at these sites, although single B. mauritanicus and B. heckerae individuals were detected. As previous work had suggested that methanotrophic and thiotrophic interactions can both occur at a site, and within an individual mussel, we investigated the symbiont communities in gill tissues of a subset of mussels from BCS and NCS. We constructed metabarcode libraries with four different primer sets spanning the 16S gene. A methanotrophic phylotype dominated all gill microbial samples from BCS, but sulfur-oxidizing Campylobacterota were represented by a notable minority of sequences from NCS. The methanotroph phylotype shared a clade with globally distributed Bathymodiolus spp. symbionts from methane seeps and hydrothermal vents. Two distinct Campylobacterota phylotypes were prevalent in NCS samples, one of which shares a clade with Campylobacterota associated with B. childressi from the Gulf of Mexico and the other with Campylobacterota associated with other deep-sea fauna. Variation in chemosynthetic symbiont communities among sites and individuals has important ecological and geochemical implications and suggests shifting reliance on methanotrophy. Continued characterization of symbionts from cold seeps will provide a greater understanding of the ecology of these unique environments as well and their geochemical footprint in elemental cycling and energy flux.
Conflict of interest statement
CSA Ocean Sciences, Inc was the management entity, and our data sharing policies were dictated by the funders – which were federal, and therefore had open data sharing. This does not alter our adherence to PLOS ONE policies on sharing data and material.
Figures




Similar articles
-
Dual symbiosis in a Bathymodiolus sp. mussel from a methane seep on the Gabon continental margin (Southeast Atlantic): 16S rRNA phylogeny and distribution of the symbionts in gills.Appl Environ Microbiol. 2005 Apr;71(4):1694-700. doi: 10.1128/AEM.71.4.1694-1700.2005. Appl Environ Microbiol. 2005. PMID: 15811991 Free PMC article.
-
Diversity, relative abundance and metabolic potential of bacterial endosymbionts in three Bathymodiolus mussel species from cold seeps in the Gulf of Mexico.Environ Microbiol. 2007 Jun;9(6):1423-38. doi: 10.1111/j.1462-2920.2007.01259.x. Environ Microbiol. 2007. PMID: 17504480
-
Characterization of tissue-associated bacterial community of two Bathymodiolus species from the adjacent cold seep and hydrothermal vent environments.Sci Total Environ. 2021 Nov 20;796:149046. doi: 10.1016/j.scitotenv.2021.149046. Epub 2021 Jul 15. Sci Total Environ. 2021. PMID: 34328889
-
Symbioses between deep-sea mussels (Mytilidae: Bathymodiolinae) and chemosynthetic bacteria: diversity, function and evolution.C R Biol. 2009 Feb-Mar;332(2-3):298-310. doi: 10.1016/j.crvi.2008.08.003. Epub 2008 Nov 25. C R Biol. 2009. PMID: 19281960 Review.
-
Macro-ecology of Gulf of Mexico cold seeps.Ann Rev Mar Sci. 2009;1:143-68. doi: 10.1146/annurev.marine.010908.163912. Ann Rev Mar Sci. 2009. PMID: 21141033 Review.
Cited by
-
The Diversity, Composition, and Putative Functions of Gill-Associated Bacteria of Bathymodiolin Mussel and Vesicomyid Clam from Haima Cold Seep, South China Sea.Microorganisms. 2020 Oct 30;8(11):1699. doi: 10.3390/microorganisms8111699. Microorganisms. 2020. PMID: 33143295 Free PMC article.
-
Composition and distribution of fish environmental DNA in an Adirondack watershed.PeerJ. 2021 Feb 26;9:e10539. doi: 10.7717/peerj.10539. eCollection 2021. PeerJ. 2021. PMID: 33680576 Free PMC article.
-
Population structure of Desmophyllum pertusum found along the United States eastern continental margin.BMC Res Notes. 2024 Oct 29;17(1):326. doi: 10.1186/s13104-024-06977-4. BMC Res Notes. 2024. PMID: 39472927 Free PMC article.
-
Genetic diversity and connectivity of chemosynthetic cold seep mussels from the U.S. Atlantic margin.BMC Ecol Evol. 2022 Jun 17;22(1):76. doi: 10.1186/s12862-022-02027-4. BMC Ecol Evol. 2022. PMID: 35715723 Free PMC article.
References
-
- Van Dover CL. The Ecology of Deep-Sea Hydrothermal Vents Princeton, NJ: Princeton University Press; 2000.
-
- Smith CR, Kukert H, Wheatcroft RA, Jumars PA, Deming JW. Vent fauna on whale remains. Nature. 1989; 341(6237):27–8. 10.1038/341027a0 WOS:A1989AN95800039 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

