Candida albicans Morphogenesis Programs Control the Balance between Gut Commensalism and Invasive Infection
- PMID: 30870623
- PMCID: PMC6581065
- DOI: 10.1016/j.chom.2019.02.008
Candida albicans Morphogenesis Programs Control the Balance between Gut Commensalism and Invasive Infection
Abstract
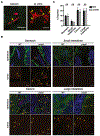
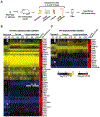
Candida albicans is a gut commensal and opportunistic pathogen. The transition between yeast and invasive hyphae is central to virulence but has unknown functions during commensal growth. In a mouse model of colonization, yeast and hyphae co-occur throughout the gastrointestinal tract. However, competitive infections of C. albicans homozygous gene disruption mutants revealed an unanticipated, inhibitory role for the yeast-to-hypha morphogenesis program on commensalism. We show that the transcription factor Ume6, a master regulator of filamentation, inhibits gut colonization, not by effects on cell shape, but by activating the expression of a hypha-specific pro-inflammatory secreted protease, Sap6, and a hyphal cell surface adhesin, Hyr1. Like a ume6 mutant, strains lacking SAP6 exhibit enhanced colonization fitness, whereas SAP6-overexpression strains are attenuated in the gut. These results reveal a tradeoff between fungal programs supporting commensalism and virulence in which selection against hypha-specific markers limits the disease-causing potential of this ubiquitous commensal-pathogen.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures




Comment in
-
Setting Up Home: Fungal Rules of Commensalism in the Mammalian Gut.Cell Host Microbe. 2019 Mar 13;25(3):347-349. doi: 10.1016/j.chom.2019.02.012. Cell Host Microbe. 2019. PMID: 30870617
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

