Comparable Genomic Copy Number Aberrations Differ across Astrocytoma Malignancy Grades
- PMID: 30871102
- PMCID: PMC6429132
- DOI: 10.3390/ijms20051251
Comparable Genomic Copy Number Aberrations Differ across Astrocytoma Malignancy Grades
Abstract
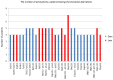
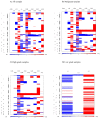
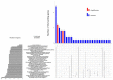
A collection of intracranial astrocytomas of different malignancy grades was analyzed for copy number aberrations (CNA) in order to identify regions that are driving cancer pathogenesis. Astrocytomas were analyzed by Array Comparative Genomic Hybridization (aCGH) and bioinformatics utilizing a Bioconductor package, Genomic Identification of Significant Targets in Cancer (GISTIC) 2.0.23 and DAVID software. Altogether, 1438 CNA were found of which losses prevailed. On our total sample, significant deletions affected 14 chromosomal regions, out of which deletions at 17p13.2, 9p21.3, 13q12.11, 22q12.3 remained significant even at 0.05 q-value. When divided into malignancy groups, the regions identified as significantly deleted in high grades were: 9p21.3; 17p13.2; 10q24.2; 14q21.3; 1p36.11 and 13q12.11, while amplified were: 3q28; 12q13.3 and 21q22.3. Low grades comprised significant deletions at 3p14.3; 11p15.4; 15q15.1; 16q22.1; 20q11.22 and 22q12.3 indicating their involvement in early stages of tumorigenesis. Significantly enriched pathways were: PI3K-Akt, Cytokine-cytokine receptor, the nucleotide-binding oligomerization domain (NOD)⁻like receptor, Jak-STAT, retinoic acid-inducible gene (RIG)-I-like receptor and Toll-like receptor pathways. HPV and herpex simplex infection and inflammation pathways were also represented. The present study brings new data to astrocytoma research amplifying the wide spectrum of changes that could help us identify the regions critical for tumorigenesis.
Keywords: GISTIC2.0.23; aCGH; astrocytoma; comparative genomic hybridization; copy number aberrations.
Conflict of interest statement
The authors declare that they have no competing interests. The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures

Similar articles
-
Detection of genetic alterations in gastric cancer patients from Saudi Arabia using comparative genomic hybridization (CGH).PLoS One. 2018 Sep 13;13(9):e0202576. doi: 10.1371/journal.pone.0202576. eCollection 2018. PLoS One. 2018. PMID: 30212456 Free PMC article.
-
Association between genomic alterations and metastatic behavior of colorectal cancer identified by array-based comparative genomic hybridization.Genes Chromosomes Cancer. 2013 Feb;52(2):140-9. doi: 10.1002/gcc.22013. Epub 2012 Oct 17. Genes Chromosomes Cancer. 2013. PMID: 23073979
-
Analysis of genomic aberrations associated with the clinicopathological parameters of rectal cancer by array‑based comparative genomic hybridization.Oncol Rep. 2013 May;29(5):1827-34. doi: 10.3892/or.2013.2296. Epub 2013 Feb 21. Oncol Rep. 2013. PMID: 23440507
-
Functionally-focused algorithmic analysis of high resolution microarray-CGH genomic landscapes demonstrates comparable genomic copy number aberrations in MSI and MSS sporadic colorectal cancer.PLoS One. 2017 Feb 23;12(2):e0171690. doi: 10.1371/journal.pone.0171690. eCollection 2017. PLoS One. 2017. PMID: 28231327 Free PMC article.
-
Microarray Techniques to Analyze Copy-Number Alterations in Genomic DNA: Array Comparative Genomic Hybridization and Single-Nucleotide Polymorphism Array.J Invest Dermatol. 2015 Oct;135(10):e37. doi: 10.1038/jid.2015.308. J Invest Dermatol. 2015. PMID: 26358390 Review. No abstract available.
Cited by
-
Searching for a signature involving 10 genes to predict the survival of patients with acute myelocytic leukemia through a combined multi-omics analysis.PeerJ. 2020 Jun 25;8:e9437. doi: 10.7717/peerj.9437. eCollection 2020. PeerJ. 2020. PMID: 32617195 Free PMC article.
-
Screening and Identification of Differentially Expressed Genes Expressed among Left and Right Colon Adenocarcinoma.Biomed Res Int. 2020 Jan 21;2020:8465068. doi: 10.1155/2020/8465068. eCollection 2020. Biomed Res Int. 2020. PMID: 32420374 Free PMC article.
-
Comprehensive analysis of single cell and bulk data develops a promising prognostic signature for improving immunotherapy responses in ovarian cancer.PLoS One. 2024 Feb 12;19(2):e0298125. doi: 10.1371/journal.pone.0298125. eCollection 2024. PLoS One. 2024. PMID: 38346070 Free PMC article.
-
Comprehensive analysis of a long non-coding RNA-associated competing endogenous RNA network in glioma.Oncol Lett. 2020 Oct;20(4):63. doi: 10.3892/ol.2020.11924. Epub 2020 Jul 29. Oncol Lett. 2020. PMID: 32863896 Free PMC article.
-
Integrative cBioPortal Analysis Revealed Molecular Mechanisms That Regulate EGFR-PI3K-AKT-mTOR Pathway in Diffuse Gliomas of the Brain.Cancers (Basel). 2021 Jun 29;13(13):3247. doi: 10.3390/cancers13133247. Cancers (Basel). 2021. PMID: 34209611 Free PMC article.
References
-
- Abou-El-Ardat K., Seifert M., Becker K., Eisenreich S., Lehmann M., Hackmann K., Rump A., Meijer G., Carvalho B., Temme A., et al. Comprehensive molecular characterization of multifocal glioblastoma proves its monoclonal origin and reveals novel insights into clonal evolution and heterogeneity of glioblastomas. Neuro-Oncology. 2017;19:546–557. doi: 10.1093/neuonc/now231. - DOI - PMC - PubMed
-
- Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

