Thinking Outside the Bug: Molecular Targets and Strategies to Overcome Antibiotic Resistance
- PMID: 30871132
- PMCID: PMC6470534
- DOI: 10.3390/ijms20061255
Thinking Outside the Bug: Molecular Targets and Strategies to Overcome Antibiotic Resistance
Abstract
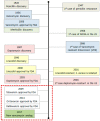
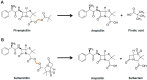
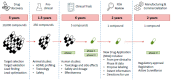
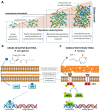
Since their discovery in the early 20th century, antibiotics have been used as the primary weapon against bacterial infections. Due to their prophylactic effect, they are also used as part of the cocktail of drugs given to treat complex diseases such as cancer or during surgery, in order to prevent infection. This has resulted in a decrease of mortality from infectious diseases and an increase in life expectancy in the last 100 years. However, as a consequence of administering antibiotics broadly to the population and sometimes misusing them, antibiotic-resistant bacteria have appeared. The emergence of resistant strains is a global health threat to humanity. Highly-resistant bacteria like Staphylococcus aureus (methicillin-resistant) or Enterococcus faecium (vancomycin-resistant) have led to complications in intensive care units, increasing medical costs and putting patient lives at risk. The appearance of these resistant strains together with the difficulty in finding new antimicrobials has alarmed the scientific community. Most of the strategies currently employed to develop new antibiotics point towards novel approaches for drug design based on prodrugs or rational design of new molecules. However, targeting crucial bacterial processes by these means will keep creating evolutionary pressure towards drug resistance. In this review, we discuss antibiotic resistance and new options for antibiotic discovery, focusing in particular on new alternatives aiming to disarm the bacteria or empower the host to avoid disease onset.
Keywords: antibiotic resistance; antimicrobial peptides and proteins; bacterial effectors; host–pathogen interactions; protein–protein interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Fleming A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzæ. Br. J. Exp. Pathol. 1929;10:226–236. doi: 10.1093/clinids/2.1.129. - DOI
-
- Rammelkamp C.H., Maxon T. Resistance of Staphylococcus aureus to the action of penicillin. Proc. Soc. Exp. Biol. Med. 1942;51:386–389. doi: 10.3181/00379727-51-13986. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

