Key Maize Drought-Responsive Genes and Pathways Revealed by Comparative Transcriptome and Physiological Analyses of Contrasting Inbred Lines
- PMID: 30871211
- PMCID: PMC6470692
- DOI: 10.3390/ijms20061268
Key Maize Drought-Responsive Genes and Pathways Revealed by Comparative Transcriptome and Physiological Analyses of Contrasting Inbred Lines
Abstract
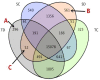
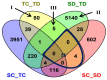
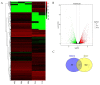
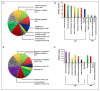
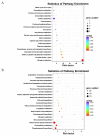
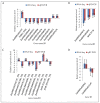
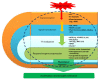
To unravel the molecular mechanisms underpinning maize (Zea mays L.) drought stress tolerance, we conducted comprehensive comparative transcriptome and physiological analyses of drought-tolerant YE8112 and drought-sensitive MO17 inbred line seedlings that had been exposed to drought treatment for seven days. Resultantly, YE8112 seedlings maintained comparatively higher leaf relative water and proline contents, greatly increased peroxidase activity, but decreased malondialdehyde content, than MO17 seedlings. Using an RNA sequencing (RNA-seq)-based approach, we identified a total of 10,612 differentially expressed genes (DEGs). From these, we mined out four critical sets of drought responsive DEGs, including 80 specific to YE8112, 5140 shared between the two lines after drought treatment (SD_TD), five DEGs of YE8112 also regulated in SD_TD, and four overlapping DEGs between the two lines. Drought-stressed YE8112 DEGs were primarily associated with nitrogen metabolism and amino-acid biosynthesis pathways, whereas MO17 DEGs were enriched in the ribosome pathway. Additionally, our physiological analyses results were consistent with the predicted RNA-seq-based findings. Furthermore, quantitative real-time polymerase chain reaction (qRT-PCR) analysis and the RNA-seq results of twenty representative DEGs were highly correlated (R² = 98.86%). Crucially, tolerant line YE8112 drought-responsive genes were predominantly implicated in stress signal transduction; cellular redox homeostasis maintenance; MYB, NAC, WRKY, and PLATZ transcriptional factor modulated; carbohydrate synthesis and cell-wall remodeling; amino acid biosynthesis; and protein ubiquitination processes. Our findings offer insights into the molecular networks mediating maize drought stress tolerance.
Keywords: RNA sequencing (RNA-seq); Zea mays L.; differentially expressed genes (DEGs); drought stress; qRT-PCR; transcriptome.
Conflict of interest statement
The authors declare that they have no conflicts of interest. Furthermore, the founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures


Similar articles
-
Comparative Proteomic and Physiological Analyses of Two Divergent Maize Inbred Lines Provide More Insights into Drought-Stress Tolerance Mechanisms.Int J Mol Sci. 2018 Oct 18;19(10):3225. doi: 10.3390/ijms19103225. Int J Mol Sci. 2018. PMID: 30340410 Free PMC article.
-
Comparative transcriptomic and physiological analyses of contrasting hybrid cultivars ND476 and ZX978 identify important differentially expressed genes and pathways regulating drought stress tolerance in maize.Genes Genomics. 2020 Aug;42(8):937-955. doi: 10.1007/s13258-020-00962-4. Epub 2020 Jul 4. Genes Genomics. 2020. PMID: 32623576
-
Comparative Proteomics and Physiological Analyses Reveal Important Maize Filling-Kernel Drought-Responsive Genes and Metabolic Pathways.Int J Mol Sci. 2019 Jul 31;20(15):3743. doi: 10.3390/ijms20153743. Int J Mol Sci. 2019. PMID: 31370198 Free PMC article.
-
Transcriptional regulatory networks in response to drought stress and rewatering in maize (Zea mays L.).Mol Genet Genomics. 2021 Nov;296(6):1203-1219. doi: 10.1007/s00438-021-01820-y. Epub 2021 Oct 3. Mol Genet Genomics. 2021. PMID: 34601650 Review.
-
Transcriptome expression profiles reveal response mechanisms to drought and drought-stress mitigation mechanisms by exogenous glycine betaine in maize.Biotechnol Lett. 2022 Mar;44(3):367-386. doi: 10.1007/s10529-022-03221-6. Epub 2022 Mar 16. Biotechnol Lett. 2022. PMID: 35294695 Review.
Cited by
-
Conventional and Omics Approaches for Understanding the Abiotic Stress Response in Cereal Crops-An Updated Overview.Plants (Basel). 2022 Oct 26;11(21):2852. doi: 10.3390/plants11212852. Plants (Basel). 2022. PMID: 36365305 Free PMC article. Review.
-
Identification of drought tolerant mechanisms in a drought-tolerant maize mutant based on physiological, biochemical and transcriptomic analyses.BMC Plant Biol. 2020 Jul 3;20(1):315. doi: 10.1186/s12870-020-02526-w. BMC Plant Biol. 2020. PMID: 32620139 Free PMC article.
-
Brassinosteroids induced drought resistance of contrasting drought-responsive genotypes of maize at physiological and transcriptomic levels.Front Plant Sci. 2022 Oct 25;13:961680. doi: 10.3389/fpls.2022.961680. eCollection 2022. Front Plant Sci. 2022. PMID: 36388543 Free PMC article.
-
Physiological and Transcriptional Analysis Reveals the Response Mechanism of Camellia vietnamensis Huang to Drought Stress.Int J Mol Sci. 2022 Oct 5;23(19):11801. doi: 10.3390/ijms231911801. Int J Mol Sci. 2022. PMID: 36233104 Free PMC article.
-
GhiPLATZ17 and GhiPLATZ22, zinc-dependent DNA-binding transcription factors, promote salt tolerance in upland cotton.Plant Cell Rep. 2024 May 13;43(6):140. doi: 10.1007/s00299-024-03178-y. Plant Cell Rep. 2024. PMID: 38740586
References
-
- Feller U., Vaseva I.I. Extreme climatic events: Impacts of drought and high temperature on physiological processes in agronomically important plants. Front. Environ. Sci. 2014;2:39. doi: 10.3389/fenvs.2014.00039. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

