Examination of Independent Prognostic Power of Gene Expressions and Histopathological Imaging Features in Cancer
- PMID: 30871256
- PMCID: PMC6468814
- DOI: 10.3390/cancers11030361
Examination of Independent Prognostic Power of Gene Expressions and Histopathological Imaging Features in Cancer
Abstract
Cancer prognosis is of essential interest, and extensive research has been conducted searching for biomarkers with prognostic power. Recent studies have shown that both omics profiles and histopathological imaging features have prognostic power. There are also studies exploring integrating the two types of measurements for prognosis modeling. However, there is a lack of study rigorously examining whether omics measurements have independent prognostic power conditional on histopathological imaging features, and vice versa. In this article, we adopt a rigorous statistical testing framework and test whether an individual gene expression measurement can improve prognosis modeling conditional on high-dimensional imaging features, and a parallel analysis is conducted reversing the roles of gene expressions and imaging features. In the analysis of The Cancer Genome Atlas (TCGA) lung adenocarcinoma and liver hepatocellular carcinoma data, it is found that multiple individual genes, conditional on imaging features, can lead to significant improvement in prognosis modeling; however, individual imaging features, conditional on gene expressions, only offer limited prognostic power. Being among the first to examine the independent prognostic power, this study may assist better understanding the "connectedness" between omics profiles and histopathological imaging features and provide important insights for data integration in cancer modeling.
Keywords: cancer prognosis; histopathological imaging features; independent prognostic power; omics profiles.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

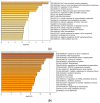
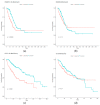
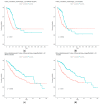
References
-
- Rath M.G., Uhlmann L., Fiedler M., Heil J., Golatta M., Dinkic C., Hennigs A., Schott S., Ernst V., Koch T., et al. Oncotype DX((R)) in breast cancer patients: Clinical experience, outcome and follow-up-a case-control study. Arch. Gynecol. Obstet. 2018;297:443–447. doi: 10.1007/s00404-017-4618-z. - DOI - PubMed
-
- Muller H.M., Widschwendter A., Fiegl H., Ivarsson L., Goebel G., Perkmann E., Marth C., Widschwendter M. DNA methylation in serum of breast cancer patients: An independent prognostic marker. Cancer Res. 2003;63:7641–7645. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

