Extended insight into the Mycobacterium chelonae-abscessus complex through whole genome sequencing of Mycobacterium salmoniphilum outbreak and Mycobacterium salmoniphilum-like strains
- PMID: 30872669
- PMCID: PMC6418233
- DOI: 10.1038/s41598-019-40922-x
Extended insight into the Mycobacterium chelonae-abscessus complex through whole genome sequencing of Mycobacterium salmoniphilum outbreak and Mycobacterium salmoniphilum-like strains
Abstract
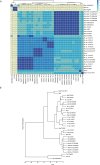
Members of the Mycobacterium chelonae-abscessus complex (MCAC) are close to the mycobacterial ancestor and includes both human, animal and fish pathogens. We present the genomes of 14 members of this complex: the complete genomes of Mycobacterium salmoniphilum and Mycobacterium chelonae type strains, seven M. salmoniphilum isolates, and five M. salmoniphilum-like strains including strains isolated during an outbreak in an animal facility at Uppsala University. Average nucleotide identity (ANI) analysis and core gene phylogeny revealed that the M. salmoniphilum-like strains are variants of the human pathogen Mycobacterium franklinii and phylogenetically close to Mycobacterium abscessus. Our data further suggested that M. salmoniphilum separates into three branches named group I, II and III with the M. salmoniphilum type strain belonging to group II. Among predicted virulence factors, the presence of phospholipase C (plcC), which is a major virulence factor that makes M. abscessus highly cytotoxic to mouse macrophages, and that M. franklinii originally was isolated from infected humans make it plausible that the outbreak in the animal facility was caused by a M. salmoniphilum-like strain. Interestingly, M. salmoniphilum-like was isolated from tap water suggesting that it can be present in the environment. Moreover, we predicted the presence of mutational hotspots in the M. salmoniphilum isolates and 26% of these hotspots overlap with genes categorized as having roles in virulence, disease and defense. We also provide data about key genes involved in transcription and translation such as sigma factor, ribosomal protein and tRNA genes.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Bataillon E, Dubard TL. Un nouveau type de tuberculose. Comptes rendus des Sceances de la Societe Biologie. 1897;49:446–449.
-
- Austin, B. & Austin, D. A. Bacterial Fish Pathogens (SpringerDordrecht Heidelberg New York London) (2013).
-
- Whitman, W. B. et al. Bergey’s Manual® of Systematic Bacteriology. 2nd ed. Springer New York: New York, NY (2012).
-
- Righetti M, et al. Mycobacterium salmoniphilum infection in a farmed Russian sturgeon, Acipenser gueldenstaedtii (Brandt & Ratzeburg) J Fish Dis. 2014;37:671–674. - PubMed
-
- Zerihun MA, Nilsen H, Hodneland S, Colquhoun DJ. Mycobacterium salmoniphilum infection in farmed Atlantic salmon, Salmo salar L. J Fish Dis. 2011;34:769–781. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

