Transcriptomic and lipidomic analysis of an EPA-containing Nannochloropsis sp. PJ12 in response to nitrogen deprivation
- PMID: 30872742
- PMCID: PMC6418175
- DOI: 10.1038/s41598-019-41169-2
Transcriptomic and lipidomic analysis of an EPA-containing Nannochloropsis sp. PJ12 in response to nitrogen deprivation
Abstract
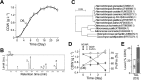
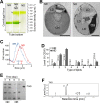
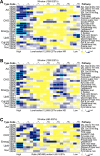
To understand genes involved in neutral lipid accumulation upon nitrogen deprivation (ND) in a novel isolate of Nannochloropsis sp. PJ12, we performed comparative transcriptomic and lipidomic analyses of cells under ND and NR (nitrogen replete) conditions. Transcriptomic profiling indicated that, while enzymes involved in TCA cycle in PJ12 under ND condition were upregulated compared to that under NR condition, those involved in Calvin cycle and glycolysis under ND condition were downregulated. Furthermore, we showed that enzymes involved in fatty acid synthesis and glycerolipid synthesis were downregulated but not β-oxidation. Lipidomic profiling indicated that, while the level of neutral lipids in ND cells was increased compared to that of NR cells, level of photosynthetic membrane-lipids DGDG and PG was decreased. Taken together, our analysis indicated that TAG accumulation is attributed to the modification of membrane lipids derived primarily from "prokaryotic" pathway and secondarily from "eukaryotic" pathway based on the 16:X or 18:X fatty acid at the sn2 position of the glycerol backbone. We propose that two-phase (NR-ND) growth is ideal for biomass and biofuel production because ND reduces cell growth rate due to the loss of photosynthetic membrane and decreased quantum yield.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Sukenik A, Carmeli Y, Berner T. Regulation of fatty acid composition by irradiance level in the Eustigmatophyte Nannochloropsis sp. Journal of Phycology. 1989;25:686–692. doi: 10.1111/j.0022-3646.1989.00686.x. - DOI
-
- Boussiba S, Vonshak A, Cohen Z, Avissar Y, Richmond A. Lipid and biomass production by the halotolerant microalga Nannochloropsis salina. Biomass. 1987;12:37–47. doi: 10.1016/0144-4565(87)90006-0. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

