Extrachromosomal oncogene amplification in tumour pathogenesis and evolution
- PMID: 30872802
- PMCID: PMC7168519
- DOI: 10.1038/s41568-019-0128-6
Extrachromosomal oncogene amplification in tumour pathogenesis and evolution
Abstract
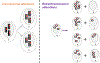
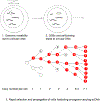
Recent reports have demonstrated that oncogene amplification on extrachromosomal DNA (ecDNA) is a frequent event in cancer, providing new momentum to explore a phenomenon first discovered several decades ago. The direct consequence of ecDNA gains in these cases is an increase in DNA copy number of the oncogenes residing on the extrachromosomal element. A secondary effect, perhaps even more important, is that the unequal segregation of ecDNA from a parental tumour cell to offspring cells rapidly increases tumour heterogeneity, thus providing the tumour with an additional array of responses to microenvironment-induced and therapy-induced stress factors and perhaps providing an evolutionary advantage. This Perspectives article discusses the current knowledge and potential implications of oncogene amplification on ecDNA in cancer.
Conflict of interest statement
Competing interests
RGWV, VB, and PSM are co-founders of and have equity interest in Pretzel Therapeutics, Inc. (PT). PSM serves as a consultant to PT. VB is a co-founder, and has equity interest in Digital Proteomics, LLC (DP), and receives income from DP. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. PT and DP were not involved in the research presented here.
Figures
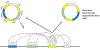
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

