Identification of Key Genes and Pathways in Post-traumatic Stress Disorder Using Microarray Analysis
- PMID: 30873067
- PMCID: PMC6403462
- DOI: 10.3389/fpsyg.2019.00302
Identification of Key Genes and Pathways in Post-traumatic Stress Disorder Using Microarray Analysis
Abstract
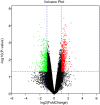
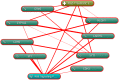
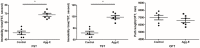
Introduction: Post-traumatic stress disorder (PTSD) is characterized by impaired fear extinction, excessive anxiety, and depression. However, the potential pathogenesis and cause of PTSD are not fully understood. Hence, the purpose of this study was to identify key genes and pathway involved in PTSD and reveal underlying molecular mechanisms by using bioinformatics analysis. Methods: The mRNA microarray expression profile dataset was retrieved and downloaded from the Gene Expression Omnibus (GEO) database. The differentially expressed genes (DEGs) were screened using GEO2R. Gene ontology (GO) was used for gene function annotations and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway was performed for enrichment analysis. Subsequently, protein-protein interaction (PPI) network and module analysis by the plugin MCODE were mapped by Cytoscape software. Finally, these key genes were verified in stress-exposed models by Real-Time quantitative (qRT-PCR). In addition, we performed text mining among the key genes and pathway with PTSD by using COREMINE. Results: A total of 1004 DEGs were identified. Gene functional annotations and enrichment analysis indicated that the most associated pathway was closely related to the Wnt signaling pathway. Using PPI network and module analysis, we identified a group of "seed" genes. These genes were further verified by qRT-PCR. In addition, text mining indicated that the altered CYP1A2, SYT1, and NLGN1 affecting PTSD might work via the Wnt signaling pathway. Conclusion: By using bioinformatics analysis, we identified a number of genes and relevant pathway which may represent key mechanisms associated with PTSD. However, these findings require verification in future experimental studies.
Keywords: PTSD; bioinformatics analysis; key genes; key pathways; microarray analysis.
Figures


Similar articles
-
Identification of Critical Signature in Post-Traumatic Stress Disorder Using Bioinformatics Analysis and in Vitro Analyses.Brain Behav. 2025 Jan;15(1):e70243. doi: 10.1002/brb3.70243. Brain Behav. 2025. PMID: 39829117 Free PMC article.
-
Identification of key genes involved in post-traumatic stress disorder: Evidence from bioinformatics analysis.World J Psychiatry. 2020 Dec 19;10(12):286-298. doi: 10.5498/wjp.v10.i12.286. eCollection 2020 Dec 19. World J Psychiatry. 2020. PMID: 33392005 Free PMC article.
-
The identification of key genes and pathways in hepatocellular carcinoma by bioinformatics analysis of high-throughput data.Med Oncol. 2017 Jun;34(6):101. doi: 10.1007/s12032-017-0963-9. Epub 2017 Apr 21. Med Oncol. 2017. PMID: 28432618 Free PMC article.
-
Identification of Gene Changes Induced by Dexamethasone in the Anterior Segment of the Human Eye Using Bioinformatics Analysis.Med Sci Monit. 2019 Jul 24;25:5501-5509. doi: 10.12659/MSM.915591. Med Sci Monit. 2019. PMID: 31339875 Free PMC article.
-
Identification of key genes and pathways in pelvic organ prolapse based on gene expression profiling by bioinformatics analysis.Arch Gynecol Obstet. 2018 May;297(5):1323-1332. doi: 10.1007/s00404-018-4745-1. Epub 2018 Mar 15. Arch Gynecol Obstet. 2018. PMID: 29546564
Cited by
-
Mitochondrial dysfunction as a possible trigger of neuroinflammation at post-traumatic stress disorder (PTSD).Front Physiol. 2023 Oct 24;14:1222826. doi: 10.3389/fphys.2023.1222826. eCollection 2023. Front Physiol. 2023. PMID: 37942228 Free PMC article. Review.
-
Increased H3K4me3 methylation and decreased miR-7113-5p expression lead to enhanced Wnt/β-catenin signaling in immune cells from PTSD patients leading to inflammatory phenotype.Mol Med. 2020 Nov 14;26(1):110. doi: 10.1186/s10020-020-00238-3. Mol Med. 2020. PMID: 33189141 Free PMC article.
-
Identification of Key Genes and the Pathophysiology Associated With Major Depressive Disorder Patients Based on Integrated Bioinformatics Analysis.Front Psychiatry. 2020 Apr 3;11:192. doi: 10.3389/fpsyt.2020.00192. eCollection 2020. Front Psychiatry. 2020. PMID: 32317989 Free PMC article.
-
Age-Dependent Effects of Chronic Stress on Zebrafish Behavior and Regeneration.Front Physiol. 2022 Apr 29;13:856778. doi: 10.3389/fphys.2022.856778. eCollection 2022. Front Physiol. 2022. PMID: 35574490 Free PMC article.
-
Genome-wide study of key genes and scoring system as potential noninvasive biomarkers for detection of suicide behavior in major depression disorder.Bioengineered. 2020 Dec;11(1):1189-1196. doi: 10.1080/21655979.2020.1831349. Bioengineered. 2020. PMID: 33103556 Free PMC article.
References
-
- Chen C., Ji M., Xu Q., Zhang Y., Sun Q., Liu J., et al. (2015). Sevoflurane attenuates stress-enhanced fear learning by regulating hippocampal BDNF expression and Akt/GSK-3beta signaling pathway in a rat model of post-traumatic stress disorder. J. Anesth. 29 600–608. 10.1007/s00540-014-1964-x - DOI - PubMed
-
- Dennis P. A., Weinberg J. B., Calhoun P. S., Watkins L. L., Sherwood A., Dennis M. F., et al. (2016). An investigation of vago-regulatory and health-behavior accounts for increased inflammation in posttraumatic stress disorder. J. Psychosom. Res. 83 33–39. 10.1016/j.jpsychores.2016.02.008 - DOI - PMC - PubMed
-
- Duan Z., He M., Zhang J., Chen K., Li B., Wang J., et al. (2015). Assessment of functional tag single nucleotide polymorphisms within the DRD2 gene as risk factors for post-traumatic stress disorder in the Han Chinese population. J. Affect. Disord. 188 210–217. 10.1016/j.jad.2015.08.066 - DOI - PubMed
LinkOut - more resources
Full Text Sources

