Cell Wall Hydrolases in Bacteria: Insight on the Diversity of Cell Wall Amidases, Glycosidases and Peptidases Toward Peptidoglycan
- PMID: 30873139
- PMCID: PMC6403190
- DOI: 10.3389/fmicb.2019.00331
Cell Wall Hydrolases in Bacteria: Insight on the Diversity of Cell Wall Amidases, Glycosidases and Peptidases Toward Peptidoglycan
Abstract
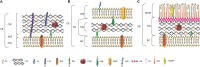
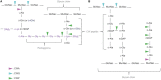
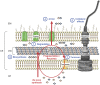
The cell wall (CW) of bacteria is an intricate arrangement of macromolecules, at least constituted of peptidoglycan (PG) but also of (lipo)teichoic acids, various polysaccharides, polyglutamate and/or proteins. During bacterial growth and division, there is a constant balance between CW degradation and biosynthesis. The CW is remodeled by bacterial hydrolases, whose activities are carefully regulated to maintain cell integrity or lead to bacterial death. Each cell wall hydrolase (CWH) has a specific role regarding the PG: (i) cell wall amidase (CWA) cleaves the amide bond between N-acetylmuramic acid and L-alanine residue at the N-terminal of the stem peptide, (ii) cell wall glycosidase (CWG) catalyses the hydrolysis of the glycosidic linkages, whereas (iii) cell wall peptidase (CWP) cleaves amide bonds between amino acids within the PG chain. After an exhaustive overview of all known conserved catalytic domains responsible for CWA, CWG, and CWP activities, this review stresses that the CWHs frequently display a modular architecture combining multiple and/or different catalytic domains, including some lytic transglycosylases as well as CW binding domains. From there, direct physiological and collateral roles of CWHs in bacterial cells are further discussed.
Keywords: bacterial cell wall; bacterial division and growth; cell lysis; cell wall binding domains; cell wall remodeling; peptidoglycan (PG) hydrolases; protein modules.
Figures




References
-
- Abaev I., Foster-Frey J., Korobova O., Shishkova N., Kiseleva N., Kopylov P., et al. . (2013). Staphylococcal Phage 2638A endolysin is lytic for Staphylococcus aureus and harbors an inter-lytic-domain secondary translational start site. Appl. Microbiol. Biotechnol. 97, 3449–3456. 10.1007/s00253-012-4252-4 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

