Extended Spectrum Beta-Lactamase-Producing Gram-Negative Bacteria Recovered From an Amazonian Lake Near the City of Belém, Brazil
- PMID: 30873145
- PMCID: PMC6403167
- DOI: 10.3389/fmicb.2019.00364
Extended Spectrum Beta-Lactamase-Producing Gram-Negative Bacteria Recovered From an Amazonian Lake Near the City of Belém, Brazil
Abstract
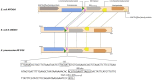
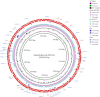
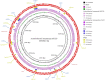
Aquatic systems have been described as antibiotic resistance reservoirs, where water may act as a vehicle for the spread of resistant bacteria and resistance genes. We evaluated the occurrence and diversity of third generation cephalosporin-resistant gram-negative bacteria in a lake in the Amazonia region. This water is used for human activities, including consumption after appropriate treatment. Eighteen samples were obtained from six sites in October 2014. Water quality parameters were generally within the legislation limits. Thirty-three bacterial isolates were identified as Escherichia (n = 7 isolates), Acinetobacter, Enterobacter, and Klebsiella (n = 5 each), Pseudomonas (n = 4), Shigella (n = 3), and Chromobacterium, Citrobacter, Leclercia, Phytobacter (1 isolate each). Twenty nine out of 33 isolates (88%) were resistant to most beta-lactams, except carbapenems, and 88% (n = 29) were resistant to antibiotics included in at least three different classes. Among the beta-lactamase genes inspected, the bla CTX-M was the most prevalent (n = 12 positive isolates), followed by bla TEM (n = 5) and bla SHV (n = 4). bla CTX-M-15 (n = 5), bla CTX-M-14 (n = 1) and bla CTX-M-2 (n = 1) variants were detected in conserved genomic contexts: bla CTX-M-15 flanked by ISEcp1 and Orf477; bla CTX-M-14 flanked by ISEcp1 and IS903; and bla CTX-M-2 associated to an ISCR element. For 4 strains the transfer of bla CTX-M was confirmed by conjugation assays. Compared with the recipient, the transconjugants showed more than 500-fold increases in the MICs of cefotaxime and 16 to 32-fold increases in the MICs of ceftazidime. Two isolates (Escherichia coli APC43A and Acinetobacter baumannii APC25) were selected for whole genome analysis. APC43A was predicted as a E. coli pathogen of the high-risk clone ST471 and serotype O154:H18. bla CTX-M-15 as well as determinants related to efflux of antibiotics, were noted in APC43A genome. A. baumannii APC25 was susceptible to carbapenems and antibiotic resistance genes detected in its genome were intrinsic determinants (e.g., bla OXA-208 and bla ADC-like). The strain was not predicted as a human pathogen and belongs to a new sequence type. Operons related to metal resistance were predicted in both genomes as well as pathogenicity and resistance islands. Results suggest a high dissemination of ESBL-producing bacteria in Lake Água Preta which, although not presenting characteristics of a strongly impacted environment, contains multi-drug resistant pathogenic strains.
Keywords: Acinetobacter baumannii; Escherichia coli; antibiotic resistance; blaCTX–M; whole genome analysis.
Figures

References
-
- Alves M. S., Pereira A., Araujo S. M., Castro B. B., Correia A. C. M., Henriques I. (2014). Seawater is a reservoir of multi-resistant Escherichia coli, including strains hosting plasmid-mediated quinolones resistance and extended-spectrum beta-lactamases genes. Front. Microbiol. 5:426. 10.3389/fmicb.2014.00426 - DOI - PMC - PubMed
-
- Ambler R. P. (1980). Structure of β-lactamases. Philos. Trans. Royal Soc. London B Biol. Sci. 289 321–331. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

