Citrus sinensis MYB Transcription Factor CsMYB85 Induce Fruit Juice Sac Lignification Through Interaction With Other CsMYB Transcription Factors
- PMID: 30873196
- PMCID: PMC6401657
- DOI: 10.3389/fpls.2019.00213
Citrus sinensis MYB Transcription Factor CsMYB85 Induce Fruit Juice Sac Lignification Through Interaction With Other CsMYB Transcription Factors
Abstract
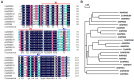
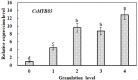
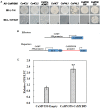
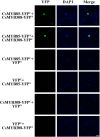
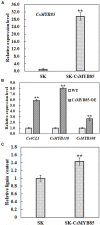
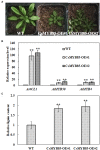
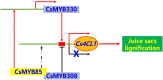
Varieties of Citrus are commercially important fruits that are cultivated worldwide and are valued for being highly nutritious and having an appealing flavor. Lignification of citrus fruit juice sacs is a serious physiological disorder that occurs during postharvest storage, for which the underlying transcriptional regulatory mechanisms remain unclear. In this study, we identified and isolated a candidate MYB transcription factor, CsMYB85, that is involved in the regulation of lignin biosynthesis in Citrus sinensis, which has homologs in Arabidopsis and other plants. We found that during juice sac lignification, CsMYB85 expression levels increase significantly, and therefore, suspected that this gene may control lignin biosynthesis during the lignification process. Our results indicated that CsMYB85 binds the CsMYB330 promoter, regulates its expression, and interacts with CsMYB308 in transgenic yeast and tobacco. A transient expression assay indicated that Cs4CL1 expression levels and lignin content significantly increased in fruit juice sacs overexpressing CsMYB85. At4CL1 expression levels and lignin content were also significantly increased in Arabidopsis overexpressing CsMYB85. We accordingly present convincing evidence for the participation of the CsMYB85 transcription factor in fruit juice sac lignification, and thereby provide new insights into the transcriptional regulation of this process in citrus fruits.
Keywords: Citrus sinensis; CsMYB85; juice sacs; lignification; postharvest.
Figures


Similar articles
-
Identification and Functional Analysis of the CgNAC043 Gene Involved in Lignin Synthesis from Citrusgrandis "San Hong".Plants (Basel). 2022 Jan 31;11(3):403. doi: 10.3390/plants11030403. Plants (Basel). 2022. PMID: 35161384 Free PMC article.
-
Citrus sinensis MYB transcription factors CsMYB330 and CsMYB308 regulate fruit juice sac lignification through fine-tuning expression of the Cs4CL1 gene.Plant Sci. 2018 Dec;277:334-343. doi: 10.1016/j.plantsci.2018.10.006. Epub 2018 Oct 10. Plant Sci. 2018. PMID: 30466599
-
Citrus NAC senescence-associated factor 1 regulates post-harvest fruit lignification via activation of 4CL1.Plant J. 2025 Aug;123(4):e70429. doi: 10.1111/tpj.70429. Plant J. 2025. PMID: 40827658
-
CsMYB15 positively regulates Cs4CL2-mediated lignin biosynthesis during juice sac granulation in navel orange.Front Plant Sci. 2023 Jun 30;14:1223820. doi: 10.3389/fpls.2023.1223820. eCollection 2023. Front Plant Sci. 2023. PMID: 37457356 Free PMC article.
-
Outgoing and potential trends of composition, health benefits, juice production and waste management of the multi-faceted Grapefruit Citrus Χ paradisi: A comprehensive review for maximizing its value.Crit Rev Food Sci Nutr. 2022;62(4):935-956. doi: 10.1080/10408398.2020.1830364. Epub 2020 Oct 15. Crit Rev Food Sci Nutr. 2022. PMID: 33054326 Review.
Cited by
-
Genome-wide identification, classification, and characterization of lectin gene superfamily in sweet orange (Citrus sinensis L.).PLoS One. 2023 Nov 13;18(11):e0294233. doi: 10.1371/journal.pone.0294233. eCollection 2023. PLoS One. 2023. PMID: 37956187 Free PMC article.
-
Identification and Functional Analysis of the CgNAC043 Gene Involved in Lignin Synthesis from Citrusgrandis "San Hong".Plants (Basel). 2022 Jan 31;11(3):403. doi: 10.3390/plants11030403. Plants (Basel). 2022. PMID: 35161384 Free PMC article.
-
Integrated Physiochemical, Hormonal, and Transcriptomic Analysis Revealed the Underlying Mechanisms for Granulation in Huyou (Citrus changshanensis) Fruit.Front Plant Sci. 2022 Jul 14;13:923443. doi: 10.3389/fpls.2022.923443. eCollection 2022. Front Plant Sci. 2022. PMID: 35909750 Free PMC article.
-
Transcriptome Analysis Unravels Metabolic and Molecular Pathways Related to Fruit Sac Granulation in a Late-Ripening Navel Orange (Citrus sinensis Osbeck).Plants (Basel). 2020 Jan 11;9(1):95. doi: 10.3390/plants9010095. Plants (Basel). 2020. PMID: 31940826 Free PMC article.
-
Effects of Efficient Ethylene Remover on the Lignification of Fresh Faba Bean (Vicia faba L.) during Storage.Foods. 2024 Sep 25;13(19):3036. doi: 10.3390/foods13193036. Foods. 2024. PMID: 39410071 Free PMC article.
References
-
- Burns J. K., Achor D. S. (1989). Cell wall changes in juice vesicles associated with “section drying” in stored late-harvested grapefruit. J. Am. Soc. Hortic. Sci. 114 283–287.
-
- Endo T., Shimada T., Fujii H., Moriguchi T., Omura M. (2007). Promoter analysis of a type 3 metallothionein-like gene abundant in Satsuma mandarin (Citrus unshiu Marc.) fruit. Sci. Hortic. 112 207–214. 10.1016/j.scienta.2006.12.042 - DOI
LinkOut - more resources
Full Text Sources

